Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells
- PMID: 25192136
- PMCID: PMC4224642
- DOI: 10.1038/nature13802
Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells
Abstract
Post-transcriptional modification of RNA nucleosides occurs in all living organisms. Pseudouridine, the most abundant modified nucleoside in non-coding RNAs, enhances the function of transfer RNA and ribosomal RNA by stabilizing the RNA structure. Messenger RNAs were not known to contain pseudouridine, but artificial pseudouridylation dramatically affects mRNA function--it changes the genetic code by facilitating non-canonical base pairing in the ribosome decoding centre. However, without evidence of naturally occurring mRNA pseudouridylation, its physiological relevance was unclear. Here we present a comprehensive analysis of pseudouridylation in Saccharomyces cerevisiae and human RNAs using Pseudo-seq, a genome-wide, single-nucleotide-resolution method for pseudouridine identification. Pseudo-seq accurately identifies known modification sites as well as many novel sites in non-coding RNAs, and reveals hundreds of pseudouridylated sites in mRNAs. Genetic analysis allowed us to assign most of the new modification sites to one of seven conserved pseudouridine synthases, Pus1-4, 6, 7 and 9. Notably, the majority of pseudouridines in mRNA are regulated in response to environmental signals, such as nutrient deprivation in yeast and serum starvation in human cells. These results suggest a mechanism for the rapid and regulated rewiring of the genetic code through inducible mRNA modifications. Our findings reveal unanticipated roles for pseudouridylation and provide a resource for identifying the targets of pseudouridine synthases implicated in human disease.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
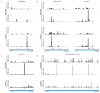

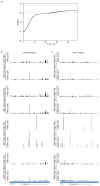
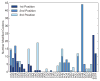
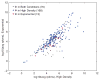
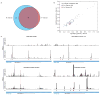


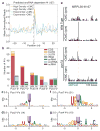
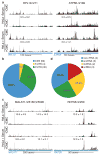
Comment in
-
Pseudouridine in a new era of RNA modifications.Cell Res. 2015 Feb;25(2):153-4. doi: 10.1038/cr.2014.143. Epub 2014 Nov 4. Cell Res. 2015. PMID: 25367125 Free PMC article.
-
Sequencing: Rereading familiar messages.Nat Methods. 2014 Nov;11(11):1095. doi: 10.1038/nmeth.3166. Nat Methods. 2014. PMID: 25551126 No abstract available.
Similar articles
-
Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae.PLoS One. 2014 Oct 29;9(10):e110799. doi: 10.1371/journal.pone.0110799. eCollection 2014. PLoS One. 2014. PMID: 25353621 Free PMC article.
-
Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA.Cell. 2014 Sep 25;159(1):148-162. doi: 10.1016/j.cell.2014.08.028. Epub 2014 Sep 11. Cell. 2014. PMID: 25219674 Free PMC article.
-
Pseudo-Seq: Genome-Wide Detection of Pseudouridine Modifications in RNA.Methods Enzymol. 2015;560:219-45. doi: 10.1016/bs.mie.2015.03.011. Epub 2015 May 28. Methods Enzymol. 2015. PMID: 26253973 Free PMC article.
-
Pseudouridylation meets next-generation sequencing.Methods. 2016 Sep 1;107:63-72. doi: 10.1016/j.ymeth.2016.03.001. Epub 2016 Mar 8. Methods. 2016. PMID: 26968262 Review.
-
Regulation and Function of RNA Pseudouridylation in Human Cells.Annu Rev Genet. 2020 Nov 23;54:309-336. doi: 10.1146/annurev-genet-112618-043830. Epub 2020 Sep 1. Annu Rev Genet. 2020. PMID: 32870730 Free PMC article. Review.
Cited by
-
Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use.Nucleic Acids Res. 2016 Sep 6;44(15):7007-78. doi: 10.1093/nar/gkw530. Epub 2016 Jul 19. Nucleic Acids Res. 2016. PMID: 27436286 Free PMC article. Review.
-
Heterogeneity of the translational machinery: Variations on a common theme.Biochimie. 2015 Jul;114:39-47. doi: 10.1016/j.biochi.2014.12.011. Epub 2014 Dec 24. Biochimie. 2015. PMID: 25542647 Free PMC article. Review.
-
Translating the epitranscriptome.Wiley Interdiscip Rev RNA. 2017 Jan;8(1):e1375. doi: 10.1002/wrna.1375. Epub 2016 Jun 27. Wiley Interdiscip Rev RNA. 2017. PMID: 27345446 Free PMC article. Review.
-
Genome-wide identification, characterization, and expression analysis unveil the roles of pseudouridine synthase (PUS) family proteins in rice development and stress response.Physiol Mol Biol Plants. 2023 Dec;29(12):1981-2004. doi: 10.1007/s12298-023-01396-4. Epub 2023 Nov 30. Physiol Mol Biol Plants. 2023. PMID: 38222285 Free PMC article.
-
RNA modifications in aging-associated cardiovascular diseases.Aging (Albany NY). 2022 Sep 29;14(19):8110-8136. doi: 10.18632/aging.204311. Epub 2022 Sep 29. Aging (Albany NY). 2022. PMID: 36178367 Free PMC article. Review.
References
-
- Davis FF, Allen FW. Ribonucleic acids from yeast which contain a fifth nucleotide. J Biol Chem. 1957;227:907–915. - PubMed
-
- Arnez JG, Steitz TA. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry. 1994;33:7560–7567. - PubMed
-
- Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. - PubMed
-
- Davis DR, Poulter CD. 1H-15N NMR studies of Escherichia coli tRNA(Phe) from hisT mutants: a structural role for pseudouridine. Biochemistry. 1991;30:4223–4231. - PubMed
-
- Davis DR, Veltri CA, Nielsen L. An RNA model system for investigation of pseudouridine stabilization of the codon-anticodon interaction in tRNALys, tRNAHis and tRNATyr. J Biomol Struct Dyn. 1998;15:1121–1132. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

