Identification of gene expression biomarkers for predicting radiation exposure
- PMID: 25189756
- PMCID: PMC4155333
- DOI: 10.1038/srep06293
Identification of gene expression biomarkers for predicting radiation exposure
Abstract
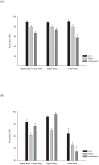
A need for more accurate and reliable radiation dosimetry has become increasingly important due to the possibility of a large-scale radiation emergency resulting from terrorism or nuclear accidents. Although traditional approaches provide accurate measurements, such methods usually require tedious effort and at least two days to complete. Therefore, we provide a new method for rapid prediction of radiation exposure. Eleven microarray datasets were classified into two groups based on their radiation doses and utilized as the training samples. For the two groups, Student's t-tests and resampling tests were used to identify biomarkers, and their gene expression ratios were used to develop a prediction model. The performance of the model was evaluated in four independent datasets, and Ingenuity pathway analysis was performed to characterize the associated biological functions. Our meta-analysis identified 29 biomarkers, showing approximately 90% and 80% accuracy in the training and validation samples. Furthermore, the 29 genes significantly participated in the regulation of cell cycle, and 19 of them are regulated by three well-known radiation-modulated transcription factors: TP53, FOXM1 and ERBB2. In conclusion, this study demonstrates a reliable method for identifying biomarkers across independent studies and high and reproducible prediction accuracy was demonstrated in both internal and external datasets.
Figures

Similar articles
-
BIOLOGICAL MARKERS OF EXTERNAL AND INTERNAL EXPOSURE IN SHELTER CONSTRUCTION WORKERS: A 13-YEAR EXPERIENCE.Radiat Prot Dosimetry. 2018 Dec 1;182(1):146-153. doi: 10.1093/rpd/ncy128. Radiat Prot Dosimetry. 2018. PMID: 30169881
-
EXPRESSION OF BIOLOGICAL MARKERS INDUCED BY IONIZING RADIATION AT THE LATE PERIOD AFTER EXPOSURE IN A WIDE RANGE OF DOSES.Probl Radiac Med Radiobiol. 2018 Dec;23:331-350. doi: 10.33145/2304-8336-2018-23-331-350. Probl Radiac Med Radiobiol. 2018. PMID: 30582855 English, Ukrainian.
-
Identification of potential radiation-responsive biomarkers based on human orthologous genes with possible roles in DNA repair pathways by comparison between Arabidopsis thaliana and homo sapiens.Sci Total Environ. 2020 Feb 1;702:135076. doi: 10.1016/j.scitotenv.2019.135076. Epub 2019 Nov 1. Sci Total Environ. 2020. PMID: 31734608
-
Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review.
-
Usefulness and limits of biological dosimetry based on cytogenetic methods.Radiat Prot Dosimetry. 2005;115(1-4):448-54. doi: 10.1093/rpd/nci061. Radiat Prot Dosimetry. 2005. PMID: 16381765 Review.
Cited by
-
Identification of marker genes in Alzheimer's disease using a machine-learning model.Bioinformation. 2021 Feb 28;17(2):348-355. doi: 10.6026/97320630017348. eCollection 2021. Bioinformation. 2021. PMID: 34234395 Free PMC article.
-
Enhanced γ-H2AX Foci Frequency and Altered Gene Expression in Participants Exposed to Ionizing Radiation During I-131 Nuclear Medicine Procedures.Nucl Med Mol Imaging. 2024 Oct;58(6):341-353. doi: 10.1007/s13139-024-00872-3. Epub 2024 Jul 12. Nucl Med Mol Imaging. 2024. PMID: 39308490
-
Transcriptional profile of immediate response to ionizing radiation exposure.Genom Data. 2015 Dec 1;7:82-5. doi: 10.1016/j.gdata.2015.11.027. eCollection 2016 Mar. Genom Data. 2015. PMID: 26981369 Free PMC article.
-
Meta-analysis approach as a gene selection method in class prediction: does it improve model performance? A case study in acute myeloid leukemia.BMC Bioinformatics. 2017 Apr 11;18(1):210. doi: 10.1186/s12859-017-1619-7. BMC Bioinformatics. 2017. PMID: 28399794 Free PMC article.
-
Biomarkers of Ionizing Radiation Exposure: A Multiparametric Approach.Genome Integr. 2017 Jan 23;8:6. doi: 10.4103/2041-9414.198911. eCollection 2017. Genome Integr. 2017. PMID: 28250913 Free PMC article.
References
-
- Ding L. H. et al. Gene expression profiles of normal human fibroblasts after exposure to ionizing radiation: a comparative study of low and high doses. Radiat Res 164, 17–26 (2005). - PubMed
-
- Short S. C. et al. Dose- and time-dependent changes in gene expression in human glioma cells after low radiation doses. Radiat Res 168, 199–208 (2007). - PubMed
-
- Donnelly E. H. et al. Acute radiation syndrome: assessment and management. South Med J 103, 541–6 (2010). - PubMed
-
- Straume T., Lucas J. N., Tucker J. D., Bigbee W. L. & Langlois R. G. Biodosimetry for a radiation worker using multiple assays. Health Phys 62, 122–30 (1992). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

