Remodeling of a delivery complex allows ClpS-mediated degradation of N-degron substrates
- PMID: 25187555
- PMCID: PMC4169940
- DOI: 10.1073/pnas.1414933111
Remodeling of a delivery complex allows ClpS-mediated degradation of N-degron substrates
Abstract
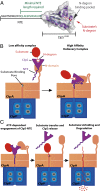
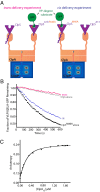
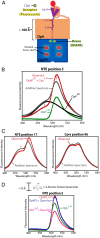
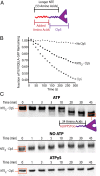
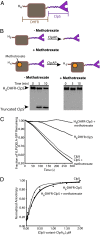
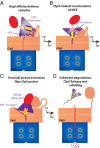
The ClpS adaptor collaborates with the AAA+ ClpAP protease to recognize and degrade N-degron substrates. ClpS binds the substrate N-degron and assembles into a high-affinity ClpS-substrate-ClpA complex, but how the N-degron is transferred from ClpS to the axial pore of the AAA+ ClpA unfoldase to initiate degradation is not known. Here we demonstrate that the unstructured N-terminal extension (NTE) of ClpS enters the ClpA processing pore in the active ternary complex. We establish that ClpS promotes delivery only in cis, as demonstrated by mixing ClpS variants with distinct substrate specificity and either active or inactive NTE truncations. Importantly, we find that ClpA engagement of the ClpS NTE is crucial for ClpS-mediated substrate delivery by using ClpS variants carrying "blocking" elements that prevent the NTE from entering the pore. These results support models in which enzymatic activity of ClpA actively remodels ClpS to promote substrate transfer, and highlight how ATPase/motor activities of AAA+ proteases can be critical for substrate selection as well as protein degradation.
Keywords: AAA+ ATPase adaptor; AAA+ unfoldase/translocase; N-degron substrate selection; adaptor remodeling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.Mol Cell. 2011 Jul 22;43(2):217-28. doi: 10.1016/j.molcel.2011.06.009. Mol Cell. 2011. PMID: 21777811 Free PMC article.
-
The Intrinsically Disordered N-terminal Extension of the ClpS Adaptor Reprograms Its Partner AAA+ ClpAP Protease.J Mol Biol. 2020 Aug 7;432(17):4908-4921. doi: 10.1016/j.jmb.2020.07.007. Epub 2020 Jul 17. J Mol Biol. 2020. PMID: 32687854 Free PMC article.
-
Division of labor between the pore-1 loops of the D1 and D2 AAA+ rings coordinates substrate selectivity of the ClpAP protease.J Biol Chem. 2021 Dec;297(6):101407. doi: 10.1016/j.jbc.2021.101407. Epub 2021 Nov 12. J Biol Chem. 2021. PMID: 34780718 Free PMC article.
-
ClpP: a structurally dynamic protease regulated by AAA+ proteins.J Struct Biol. 2012 Aug;179(2):202-10. doi: 10.1016/j.jsb.2012.05.003. Epub 2012 May 14. J Struct Biol. 2012. PMID: 22595189 Review.
-
Proteolysis: Adaptor, adaptor, catch me a catch.Curr Biol. 2004 Nov 9;14(21):R924-6. doi: 10.1016/j.cub.2004.10.015. Curr Biol. 2004. PMID: 15530384 Review.
Cited by
-
An Iterative, Synthetic Approach To Engineer a High-Performance PhoB-Specific Reporter.Appl Environ Microbiol. 2018 Jul 2;84(14):e00603-18. doi: 10.1128/AEM.00603-18. Print 2018 Jul 15. Appl Environ Microbiol. 2018. PMID: 29752265 Free PMC article.
-
Chloroplast Proteases: Updates on Proteolysis within and across Suborganellar Compartments.Plant Physiol. 2016 Aug;171(4):2280-93. doi: 10.1104/pp.16.00330. Epub 2016 Jun 10. Plant Physiol. 2016. PMID: 27288365 Free PMC article. Review.
-
The ATF3 Transcription Factor Is a Short-Lived Substrate of the Arg/N-Degron Pathway.Biochemistry. 2020 Aug 4;59(30):2796-2812. doi: 10.1021/acs.biochem.0c00514. Epub 2020 Jul 21. Biochemistry. 2020. PMID: 32692156 Free PMC article.
-
Formyl-methionine as an N-degron of a eukaryotic N-end rule pathway.Science. 2018 Nov 30;362(6418):eaat0174. doi: 10.1126/science.aat0174. Epub 2018 Nov 8. Science. 2018. PMID: 30409808 Free PMC article.
-
Formyl-methionine as a degradation signal at the N-termini of bacterial proteins.Microb Cell. 2015;2(10):376-393. doi: 10.15698/mic2015.10.231. Microb Cell. 2015. PMID: 26866044 Free PMC article.
References
-
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9(1):27–43. - PubMed
-
- Hanson PI, Whiteheart SW. AAA+ proteins: Have engine, will work. Nat Rev Mol Cell Biol. 2005;6(7):519–529. - PubMed
-
- Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. - PubMed
-
- Wah DA, Levchenko I, Baker TA, Sauer RT. Characterization of a specificity factor for an AAA+ ATPase: assembly of SspB dimers with ssrA-tagged proteins and the ClpX hexamer. Chem Biol. 2002;9(11):1237–1245. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

