RNA processing factor 7 and polynucleotide phosphorylase are necessary for processing and stability of nad2 mRNA in Arabidopsis mitochondria
- PMID: 25181358
- PMCID: PMC4179969
- DOI: 10.4161/rna.29781
RNA processing factor 7 and polynucleotide phosphorylase are necessary for processing and stability of nad2 mRNA in Arabidopsis mitochondria
Abstract
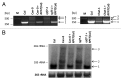
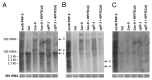
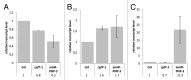
Post-transcriptional maturation of plant mitochondrial transcripts requires several steps. Among these, the generation of mature 5' ends is still one of the most enigmatic processes. Toward a characterization of proteins involved in 5' processing of mitochondrial transcripts in Arabidopsis (Arabidopsis thaliana), we now analyzed 5' maturation of nad2 transcripts. Based on natural genetic variation affecting 5' ends of nad2 transcripts in ecotype Can-0 and complementation studies we now identified RNA processing factor 7, which takes part in the generation of the 5' terminus of the mature nad2 mRNA. RPF7 is a relatively short regular P-class pentatricopeptide repeat protein comprising seven canonical P repeats and a single short S repeat. The corresponding allele in Can-0 encodes a truncated version of this protein lacking two C-terminal repeats, which are essential for the function of RPF7. Furthermore we established transgenic plants expressing artifical microRNAs targeting the mitochondrial polynucleotide phosphorylase (PNPase), which results in substantial reduction of the PNPase mRNA levels and strong knockdown of this gene. Detailed quantitative studies of 5' and 3' extended nad2 precursor RNAs in these knockdown plants as well as in the rpf7-1 knockout mutant suggest that 5' processing contributes to the stability of mitochondrial transcripts in plants.
Keywords: 5′ end processing; RNA; RNA processing factor; arabidopsis thaliana; mitochondria; pentatricopeptide repeat proteins; polynucleotide phosphorylase.
Figures


Similar articles
-
In Arabidopsis thaliana mitochondria 5' end polymorphisms of nad4L-atp4 and nad3-rps12 transcripts are linked to RNA PROCESSING FACTORs 1 and 8.Plant Mol Biol. 2021 Jul;106(4-5):335-348. doi: 10.1007/s11103-021-01153-9. Epub 2021 Apr 28. Plant Mol Biol. 2021. PMID: 33909186 Free PMC article.
-
RNA Processing Factor 5 is required for efficient 5' cleavage at a processing site conserved in RNAs of three different mitochondrial genes in Arabidopsis thaliana.Plant J. 2013 May;74(4):593-604. doi: 10.1111/tpj.12143. Epub 2013 Apr 15. Plant J. 2013. PMID: 23398165
-
Pentatricopeptide repeat protein MITOCHONDRIAL STABILITY FACTOR 3 ensures mitochondrial RNA stability and embryogenesis.Plant Physiol. 2022 Aug 29;190(1):669-681. doi: 10.1093/plphys/kiac309. Plant Physiol. 2022. PMID: 35751603 Free PMC article.
-
P-class pentatricopeptide repeat proteins are required for efficient 5' end formation of plant mitochondrial transcripts.RNA Biol. 2013;10(9):1511-9. doi: 10.4161/rna.26129. Epub 2013 Aug 15. RNA Biol. 2013. PMID: 24184847 Free PMC article. Review.
-
Maturation of 5' ends of plant mitochondrial RNAs.Physiol Plant. 2016 Jul;157(3):280-8. doi: 10.1111/ppl.12423. Epub 2016 Mar 23. Physiol Plant. 2016. PMID: 26833432 Review.
Cited by
-
The pentatricopeptide repeat protein MTSF2 stabilizes a nad1 precursor transcript and defines the 3΄ end of its 5΄-half intron.Nucleic Acids Res. 2017 Jun 2;45(10):6119-6134. doi: 10.1093/nar/gkx162. Nucleic Acids Res. 2017. PMID: 28334831 Free PMC article.
-
Mitochondrial mRNA Processing in the Chlorophyte Alga Pediastrum duplex and Streptophyte Alga Chara vulgaris Reveals an Evolutionary Branch in Mitochondrial mRNA Processing.Plants (Basel). 2021 Mar 18;10(3):576. doi: 10.3390/plants10030576. Plants (Basel). 2021. PMID: 33803683 Free PMC article.
-
Defects of mitochondrial RNA turnover lead to the accumulation of double-stranded RNA in vivo.PLoS Genet. 2019 Jul 31;15(7):e1008240. doi: 10.1371/journal.pgen.1008240. eCollection 2019 Jul. PLoS Genet. 2019. PMID: 31365523 Free PMC article.
-
In Arabidopsis thaliana distinct alleles encoding mitochondrial RNA PROCESSING FACTOR 4 support the generation of additional 5' termini of ccmB transcripts.Plant Mol Biol. 2017 Apr;93(6):659-668. doi: 10.1007/s11103-017-0591-y. Epub 2017 Feb 22. Plant Mol Biol. 2017. PMID: 28229269
-
In Arabidopsis thaliana mitochondria 5' end polymorphisms of nad4L-atp4 and nad3-rps12 transcripts are linked to RNA PROCESSING FACTORs 1 and 8.Plant Mol Biol. 2021 Jul;106(4-5):335-348. doi: 10.1007/s11103-021-01153-9. Epub 2021 Apr 28. Plant Mol Biol. 2021. PMID: 33909186 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
