Proteomics of Fusobacterium nucleatum within a model developing oral microbial community
- PMID: 25155235
- PMCID: PMC4234264
- DOI: 10.1002/mbo3.204
Proteomics of Fusobacterium nucleatum within a model developing oral microbial community
Abstract
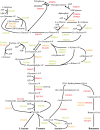
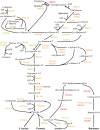
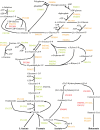
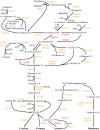
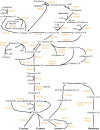
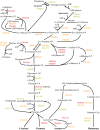
Fusobacterium nucleatum is a common oral organism that can provide adhesive and metabolic support to developing periodontal bacterial communities. It is within the context of these communities that disease occurs. We have previously reported whole cell proteomics analyses of Porphyromonas gingivalis and Streptococcus gordonii in early-stage communities with each other and with F. nucleatum, modeled using 18 h pellets. Here, we report the adaptation of F. nucleatum to the same experimental conditions as measured by differential protein expression. About 1210 F. nucleatum proteins were detected in single species F. nucleatum control samples, 1192 in communities with P. gingivalis, 1224 with S. gordonii, and 1135 with all three species. Quantitative comparisons among the proteomes revealed important changes in all mixed samples with distinct responses to P. gingivalis or S. gordonii alone and in combination. The results were inspected manually and an ontology analysis conducted using DAVID (Database for annotation, visualization, and integrated discovery). Extensive changes were detected in energy metabolism. All multispecies comparisons showed reductions in amino acid fermentation and a shift toward butanoate as a metabolic byproduct, although the two organism model community with S. gordonii showed increases in alanine, threonine, methionine, and cysteine pathways, and in the three species samples there were increases in lysine and methionine. The communities with P. gingivalis or all three organisms showed reduced glycolysis proteins, but F. nucleatum paired with S. gordonii displayed increased glycolysis/gluconeogenesis proteins. The S. gordonii containing two organism model also showed increases in the ethanolamine pathway while the three species sample showed decreases relative to the F. nucleatum single organism control. All of the nascent model communities displayed reduced translation, lipopolysaccharide, and cell wall biosynthesis, DNA replication and DNA repair.
Keywords: Biofilm; Fusobacterium nucleatum; microbial proteomics; microbiome; oral microbial ecology.
© 2014 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures

Similar articles
-
Proteomics of Streptococcus gordonii within a model developing oral microbial community.BMC Microbiol. 2012 Sep 18;12:211. doi: 10.1186/1471-2180-12-211. BMC Microbiol. 2012. PMID: 22989070 Free PMC article.
-
Label-free quantitative proteomic analysis of the oral bacteria Fusobacterium nucleatum and Porphyromonas gingivalis to identify protein features relevant in biofilm formation.Anaerobe. 2021 Dec;72:102449. doi: 10.1016/j.anaerobe.2021.102449. Epub 2021 Sep 17. Anaerobe. 2021. PMID: 34543761
-
Fusobacterium nucleatum Metabolically Integrates Commensals and Pathogens in Oral Biofilms.mSystems. 2022 Aug 30;7(4):e0017022. doi: 10.1128/msystems.00170-22. Epub 2022 Jul 19. mSystems. 2022. PMID: 35852319 Free PMC article.
-
Reexamining the role of Fusobacterium nucleatum subspecies in clinical and experimental studies.Gut Microbes. 2024 Jan-Dec;16(1):2415490. doi: 10.1080/19490976.2024.2415490. Epub 2024 Oct 12. Gut Microbes. 2024. PMID: 39394990 Free PMC article. Review.
-
Fusobacterium nucleatum - Friend or foe?J Inorg Biochem. 2021 Nov;224:111586. doi: 10.1016/j.jinorgbio.2021.111586. Epub 2021 Aug 18. J Inorg Biochem. 2021. PMID: 34425476 Review.
Cited by
-
Correlation between relative bacterial activity and lactate dehydrogenase gene expression of co-cultures in vitro.Clin Oral Investig. 2019 Mar;23(3):1225-1235. doi: 10.1007/s00784-018-2547-2. Epub 2018 Jul 6. Clin Oral Investig. 2019. PMID: 29980934
-
Fusobacterium nucleatum induces MDSCs enrichment via activation the NLRP3 inflammosome in ESCC cells, leading to cisplatin resistance.Ann Med. 2022 Dec;54(1):989-1003. doi: 10.1080/07853890.2022.2061045. Ann Med. 2022. PMID: 35435776 Free PMC article.
-
Proteomic profiling of host-biofilm interactions in an oral infection model resembling the periodontal pocket.Sci Rep. 2015 Nov 3;5:15999. doi: 10.1038/srep15999. Sci Rep. 2015. PMID: 26525412 Free PMC article.
-
Insights into Dynamic Polymicrobial Synergy Revealed by Time-Coursed RNA-Seq.Front Microbiol. 2017 Feb 28;8:261. doi: 10.3389/fmicb.2017.00261. eCollection 2017. Front Microbiol. 2017. PMID: 28293219 Free PMC article.
-
Fusobacterium nucleatum - symbiont, opportunist and oncobacterium.Nat Rev Microbiol. 2019 Mar;17(3):156-166. doi: 10.1038/s41579-018-0129-6. Nat Rev Microbiol. 2019. PMID: 30546113 Free PMC article. Review.
References
-
- Babu JP, Dean JW. Pabst MJ. Attachment of Fusobacterium nucleatum to fibronectin immobilized on gingival epithelial cells or glass coverslips. J. Periodontal. 1995;66:285–290. - PubMed
-
- Bakken V, Högh BT. Jensen HB. Utilization of amino acids and peptides by Fusobacterium nulceatum. Scand. J. Dent. Res. 1989;97:43–53. - PubMed
-
- Baumgartner JC, Falker WA., Jr Beckerman T. Experimentally induced infection by oral anaerobic microorganisms in a mouse model. Oral Microbiol. Immunol. 1992;7:253–256. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

