Somatic mutations in cerebral cortical malformations
- PMID: 25140959
- PMCID: PMC4274952
- DOI: 10.1056/NEJMoa1314432
Somatic mutations in cerebral cortical malformations
Abstract
Background: Although there is increasing recognition of the role of somatic mutations in genetic disorders, the prevalence of somatic mutations in neurodevelopmental disease and the optimal techniques to detect somatic mosaicism have not been systematically evaluated.
Methods: Using a customized panel of known and candidate genes associated with brain malformations, we applied targeted high-coverage sequencing (depth, ≥200×) to leukocyte-derived DNA samples from 158 persons with brain malformations, including the double-cortex syndrome (subcortical band heterotopia, 30 persons), polymicrogyria with megalencephaly (20), periventricular nodular heterotopia (61), and pachygyria (47). We validated candidate mutations with the use of Sanger sequencing and, for variants present at unequal read depths, subcloning followed by colony sequencing.
Results: Validated, causal mutations were found in 27 persons (17%; range, 10 to 30% for each phenotype). Mutations were somatic in 8 of the 27 (30%), predominantly in persons with the double-cortex syndrome (in whom we found mutations in DCX and LIS1), persons with periventricular nodular heterotopia (FLNA), and persons with pachygyria (TUBB2B). Of the somatic mutations we detected, 5 (63%) were undetectable with the use of traditional Sanger sequencing but were validated through subcloning and subsequent sequencing of the subcloned DNA. We found potentially causal mutations in the candidate genes DYNC1H1, KIF5C, and other kinesin genes in persons with pachygyria.
Conclusions: Targeted sequencing was found to be useful for detecting somatic mutations in patients with brain malformations. High-coverage sequencing panels provide an important complement to whole-exome and whole-genome sequencing in the evaluation of somatic mutations in neuropsychiatric disease. (Funded by the National Institute of Neurological Disorders and Stroke and others.).
Figures
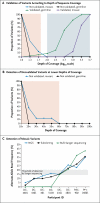
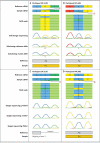
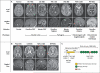
Comment in
-
Somatic mutations in cerebral cortical malformations.N Engl J Med. 2014 Nov 20;371(21):2038. doi: 10.1056/NEJMc1411784. N Engl J Med. 2014. PMID: 25409382 No abstract available.
-
Somatic mutations in cerebral cortical malformations.N Engl J Med. 2014 Nov 20;371(21):2037. doi: 10.1056/NEJMc1411784. N Engl J Med. 2014. PMID: 25409383 No abstract available.
Similar articles
-
[Epileptogenic brain malformations: radiological and clinical presentation and indications for genetic testing].Rev Neurol (Paris). 2008 Dec;164(12):995-1009. doi: 10.1016/j.neurol.2008.04.006. Epub 2008 Jun 9. Rev Neurol (Paris). 2008. PMID: 18808783 Review. French.
-
Identification of DCX gene mutation in lissencephaly spectrum with subcortical band heterotopia using whole exome sequencing.Pediatr Neurol. 2013 May;48(5):411-4. doi: 10.1016/j.pediatrneurol.2012.12.033. Pediatr Neurol. 2013. PMID: 23583063
-
Analysis of 17 genes detects mutations in 81% of 811 patients with lissencephaly.Genet Med. 2018 Nov;20(11):1354-1364. doi: 10.1038/gim.2018.8. Epub 2018 Apr 19. Genet Med. 2018. PMID: 29671837 Free PMC article.
-
Targeted re-sequencing in malformations of cortical development: genotype-phenotype correlations.Seizure. 2020 Aug;80:145-152. doi: 10.1016/j.seizure.2020.05.023. Epub 2020 Jun 3. Seizure. 2020. PMID: 32570172
-
Somatic mutation, genomic variation, and neurological disease.Science. 2013 Jul 5;341(6141):1237758. doi: 10.1126/science.1237758. Science. 2013. PMID: 23828942 Free PMC article. Review.
Cited by
-
Hemispheric cortical dysplasia secondary to a mosaic somatic mutation in MTOR.Neurology. 2015 May 19;84(20):2029-32. doi: 10.1212/WNL.0000000000001594. Epub 2015 Apr 15. Neurology. 2015. PMID: 25878179 Free PMC article.
-
Application of Peptide Nucleic Acid-based Assays Toward Detection of Somatic Mosaicism.Mol Ther Nucleic Acids. 2016 Apr 26;5(4):e314. doi: 10.1038/mtna.2016.22. Mol Ther Nucleic Acids. 2016. PMID: 27115839 Free PMC article.
-
How to proceed after "negative" exome: A review on genetic diagnostics, limitations, challenges, and emerging new multiomics techniques.J Inherit Metab Dis. 2022 Jul;45(4):663-681. doi: 10.1002/jimd.12507. Epub 2022 May 22. J Inherit Metab Dis. 2022. PMID: 35506430 Free PMC article. Review.
-
TUBB2B Mutation in an Adult Patient with Myoclonus-Dystonia.Case Rep Neurol. 2017 Aug 31;9(2):216-221. doi: 10.1159/000479788. eCollection 2017 May-Aug. Case Rep Neurol. 2017. PMID: 28966590 Free PMC article.
-
Rare copy number variations affecting the synaptic gene DMXL2 in neurodevelopmental disorders.J Neurodev Disord. 2019 Feb 7;11(1):3. doi: 10.1186/s11689-019-9263-3. J Neurodev Disord. 2019. PMID: 30732576 Free PMC article.
References
-
- Biesecker LG, Spinner NB. A genomic view of mosaicism and human disease. Nat Rev Genet. 2013;14:307–20. - PubMed
-
- Weinstein LS, Shenker A, Gejman PV, Merino MJ, Friedman E, Spiegel AM. Activating mutations of the stimulatory G protein in the McCune–Albright syndrome. N Engl J Med. 1991;325:1688–95. - PubMed
Publication types
MeSH terms
Grants and funding
- U54 HG006493/HG/NHGRI NIH HHS/United States
- RC2 MH089952/MH/NIMH NIH HHS/United States
- R01NS079277/NS/NINDS NIH HHS/United States
- 1RC2MH089952/MH/NIMH NIH HHS/United States
- R01 NS035129/NS/NINDS NIH HHS/United States
- K23NS069784/NS/NINDS NIH HHS/United States
- UM1 HG006493/HG/NHGRI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- K23 NS069784/NS/NINDS NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- T32GM07753/GM/NIGMS NIH HHS/United States
- HG006493/HG/NHGRI NIH HHS/United States
- R01NS035129/NS/NINDS NIH HHS/United States
- R01 NS079277/NS/NINDS NIH HHS/United States
- R01 NS073601/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
