Influence of Sequence and Covalent Modifications on Yeast tRNA Dynamics
- PMID: 25136272
- PMCID: PMC4132867
- DOI: 10.1021/ct500107y
Influence of Sequence and Covalent Modifications on Yeast tRNA Dynamics
Abstract
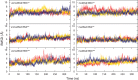
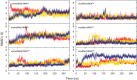
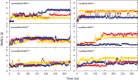
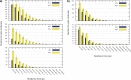
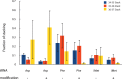
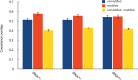
Modified nucleotides are prevalent in tRNA. Experimental studies reveal that these covalent modifications play an important role in tuning tRNA function. In this study, molecular dynamics (MD) simulations were used to investigate how modifications alter tRNA dynamics. The X-ray crystal structures of tRNA(Asp), tRNA(Phe), and tRNA(iMet), both with and without modifications, were used as initial structures for 333 ns explicit solvent MD simulations with AMBER. For each tRNA molecule, three independent trajectory calculations were performed, giving an aggregate of 6 μs of total MD across six molecules. The global root-mean-square deviations (RMSD) of atomic positions show that modifications only introduce significant rigidity to the global structure of tRNA(Phe). Interestingly, RMSDs of the anticodon stem-loop (ASL) suggest that modified tRNA has a more rigid structure compared to the unmodified tRNA in this domain. The anticodon RMSDs of the modified tRNAs, however, are higher than those of corresponding unmodified tRNAs. These findings suggest that the rigidity of the anticodon stem-loop is finely tuned by modifications, where rigidity in the anticodon arm is essential for tRNA translocation in the ribosome, and flexibility of the anticodon is important for codon recognition. Sugar pucker and water residence time of pseudouridines in modified tRNAs and corresponding uridines in unmodified tRNAs were assessed, and the results reinforce that pseudouridine favors the 3'-endo conformation and has a higher tendency to interact with water. Principal component analysis (PCA) was used to examine correlated motions in tRNA. Additionally, covariance overlaps of PCAs were compared for trajectories of the same molecule and between trajectories of modified and unmodified tRNAs. The comparison suggests that modifications alter the correlated motions. For the anticodon bases, the extent of stacking was compared between modified and unmodified molecules, and only unmodified tRNA(Asp) has significantly higher percentage of stacking time. Overall, the simulations reveal that the effect of covalent modification on tRNA dynamics is not simple, with modifications increasing flexibility in some regions of the structure and increasing rigidity in other regions.
Figures
Similar articles
-
Solution conformations of unmodified and A(37)N(6)-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNA(Phe).J Mol Biol. 2002 Jun 21;319(5):1015-34. doi: 10.1016/S0022-2836(02)00382-0. J Mol Biol. 2002. PMID: 12079344
-
Actions of the anticodon arm in translation on the phenotypes of RNA mutants.J Mol Biol. 1986 Nov 20;192(2):235-55. doi: 10.1016/0022-2836(86)90362-1. J Mol Biol. 1986. PMID: 2435916
-
Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A.Biochemistry. 2000 Nov 7;39(44):13396-404. doi: 10.1021/bi0013039. Biochemistry. 2000. PMID: 11063577
-
The effect of pseudouridine and pH on the structure and dynamics of the anticodon stem-loop of tRNA(Lys,3).Nucleic Acids Symp Ser. 1997;(36):56-7. Nucleic Acids Symp Ser. 1997. PMID: 9478205 Review.
-
The Importance of Being Modified: The Role of RNA Modifications in Translational Fidelity.Enzymes. 2017;41:1-50. doi: 10.1016/bs.enz.2017.03.005. Epub 2017 Apr 22. Enzymes. 2017. PMID: 28601219 Free PMC article. Review.
Cited by
-
Direct interplay between stereochemistry and conformational preferences in aminoacylated oligoribonucleotides.Nucleic Acids Res. 2019 Dec 2;47(21):11077-11089. doi: 10.1093/nar/gkz902. Nucleic Acids Res. 2019. PMID: 31612955 Free PMC article.
-
Structural effects of modified ribonucleotides and magnesium in transfer RNAs.Bioorg Med Chem. 2016 Oct 15;24(20):4826-4834. doi: 10.1016/j.bmc.2016.06.037. Epub 2016 Jun 18. Bioorg Med Chem. 2016. PMID: 27364608 Free PMC article.
-
Adenine Methylation Enhances the Conformational Flexibility of an RNA Hairpin Tetraloop.J Phys Chem B. 2024 Apr 4;128(13):3157-3166. doi: 10.1021/acs.jpcb.4c00522. Epub 2024 Mar 27. J Phys Chem B. 2024. PMID: 38535997 Free PMC article.
-
Challenges with Simulating Modified RNA: Insights into Role and Reciprocity of Experimental and Computational Approaches.Genes (Basel). 2022 Mar 18;13(3):540. doi: 10.3390/genes13030540. Genes (Basel). 2022. PMID: 35328093 Free PMC article. Review.
-
Protein DEK and DTA Aptamers: Insight Into the Interaction Mechanisms and the Computational Aptamer Design.Front Mol Biosci. 2022 Jul 19;9:946480. doi: 10.3389/fmolb.2022.946480. eCollection 2022. Front Mol Biosci. 2022. PMID: 35928230 Free PMC article.
References
-
- Persson B. C. Modification of tRNA as a regulatory device. Mol. Microbiol. 1993, 8, 1011–1016. - PubMed
-
- Björk G. R.; Durand J.; Hagervall T. G.; Lundgren H. K.; Nilsson K.; Chen P.; Qian Q.; Urbonavičius J. Transfer RNA modification: Influence on translational frameshifting and metabolism. FEBS Lett. 1999, 452, 47–51. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
