ExaBayes: massively parallel bayesian tree inference for the whole-genome era
- PMID: 25135941
- PMCID: PMC4166930
- DOI: 10.1093/molbev/msu236
ExaBayes: massively parallel bayesian tree inference for the whole-genome era
Abstract
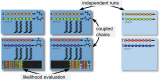
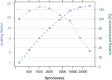
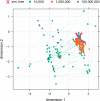
Modern sequencing technology now allows biologists to collect the entirety of molecular evidence for reconstructing evolutionary trees. We introduce a novel, user-friendly software package engineered for conducting state-of-the-art Bayesian tree inferences on data sets of arbitrary size. Our software introduces a nonblocking parallelization of Metropolis-coupled chains, modifications for efficient analyses of data sets comprising thousands of partitions and memory saving techniques. We report on first experiences with Bayesian inferences at the whole-genome level using the SuperMUC supercomputer and simulated data.
Keywords: Bayesian statistics; parallelization; phylogenetic inference; software; whole-genome analyses.
© The Author 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
Similar articles
-
Bayesian coestimation of phylogeny and sequence alignment.BMC Bioinformatics. 2005 Apr 1;6:83. doi: 10.1186/1471-2105-6-83. BMC Bioinformatics. 2005. PMID: 15804354 Free PMC article.
-
Species trees from highly incongruent gene trees in rice.Syst Biol. 2009 Oct;58(5):489-500. doi: 10.1093/sysbio/syp054. Epub 2009 Sep 21. Syst Biol. 2009. PMID: 20525603
-
Bayesian Molecular Clock Dating Using Genome-Scale Datasets.Methods Mol Biol. 2019;1910:309-330. doi: 10.1007/978-1-4939-9074-0_10. Methods Mol Biol. 2019. PMID: 31278669
-
Recent advances in computational phylodynamics.Curr Opin Virol. 2018 Aug;31:24-32. doi: 10.1016/j.coviro.2018.08.009. Epub 2018 Sep 22. Curr Opin Virol. 2018. PMID: 30248578 Review.
-
Are you my mother? Bayesian phylogenetic inference of recombination among putative parental strains.Appl Bioinformatics. 2003;2(3):131-44. Appl Bioinformatics. 2003. PMID: 15130798 Review.
Cited by
-
Cultivating epizoic diatoms provides insights into the evolution and ecology of both epibionts and hosts.Sci Rep. 2022 Sep 6;12(1):15116. doi: 10.1038/s41598-022-19064-0. Sci Rep. 2022. PMID: 36068258 Free PMC article.
-
Revisiting the hyperdominance of Neotropical tree species under a taxonomic, functional and evolutionary perspective.Sci Rep. 2021 May 5;11(1):9585. doi: 10.1038/s41598-021-88417-y. Sci Rep. 2021. PMID: 33953271 Free PMC article.
-
Fungi with history: Unveiling the mycobiota of historic documents of Costa Rica.PLoS One. 2023 Jan 18;18(1):e0279914. doi: 10.1371/journal.pone.0279914. eCollection 2023. PLoS One. 2023. PMID: 36652424 Free PMC article.
-
Parallel power posterior analyses for fast computation of marginal likelihoods in phylogenetics.PeerJ. 2021 Nov 2;9:e12438. doi: 10.7717/peerj.12438. eCollection 2021. PeerJ. 2021. PMID: 34760401 Free PMC article.
-
Detection and Characterization of Invertebrate Iridoviruses Found in Reptiles and Prey Insects in Europe over the Past Two Decades.Viruses. 2019 Jul 2;11(7):600. doi: 10.3390/v11070600. Viruses. 2019. PMID: 31269721 Free PMC article.
References
-
- Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. Parallel Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics. 2004;20(3):407–415. - PubMed
-
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452(7188):745–749. - PubMed
-
- Fitch WM, Margoliash E. Construction of phylogenetic trees. Science. 1967;155(760):279–284. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

