Targeting of viral capsids to nuclear pores in a cell-free reconstitution system
- PMID: 25131140
- PMCID: PMC5524173
- DOI: 10.1111/tra.12209
Targeting of viral capsids to nuclear pores in a cell-free reconstitution system
Abstract
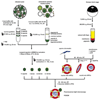
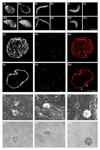
Many viruses deliver their genomes into the nucleoplasm for viral transcription and replication. Here, we describe a novel cell-free system to elucidate specific interactions between viruses and nuclear pore complexes (NPCs). Nuclei reconstituted in vitro from egg extracts of Xenopus laevis, an established biochemical system to decipher nuclear functions, were incubated with GFP-tagged capsids of herpes simplex virus, an alphaherpesvirus replicating in the nucleus. Capsid binding to NPCs was analyzed using fluorescence and field emission scanning electron microscopy. Tegument-free capsids or viral capsids exposing inner tegument proteins on their surface bound to nuclei, while capsids inactivated by a high-salt treatment or covered by inner and outer tegument showed less binding. There was little binding of the four different capsid types to nuclei lacking functional NPCs. This novel approach provides a powerful system to elucidate the molecular mechanisms that enable viral structures to engage with NPCs. Furthermore, this assay could be expanded to identify molecular cues triggering viral genome uncoating and nuclear import of viral genomes.
Keywords: Xenopus nuclei; herpes simplex virus; herpesviruses; nuclear pore; reconstitution.
© 2014 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Figures


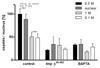
Similar articles
-
The C Terminus of the Herpes Simplex Virus UL25 Protein Is Required for Release of Viral Genomes from Capsids Bound to Nuclear Pores.J Virol. 2017 Jul 12;91(15):e00641-17. doi: 10.1128/JVI.00641-17. Print 2017 Aug 1. J Virol. 2017. PMID: 28490590 Free PMC article.
-
Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro.Mol Cell Biol. 2000 Jul;20(13):4922-31. doi: 10.1128/MCB.20.13.4922-4931.2000. Mol Cell Biol. 2000. PMID: 10848617 Free PMC article.
-
Role of HSV-1 Capsid Vertex-Specific Component (CVSC) and Viral Terminal DNA in Capsid Docking at the Nuclear Pore.Viruses. 2021 Dec 15;13(12):2515. doi: 10.3390/v13122515. Viruses. 2021. PMID: 34960783 Free PMC article.
-
A Hitchhiker's Guide Through the Cell: The World According to the Capsids of Alphaherpesviruses.Annu Rev Virol. 2024 Sep;11(1):215-238. doi: 10.1146/annurev-virology-100422-022751. Epub 2024 Aug 30. Annu Rev Virol. 2024. PMID: 38954634 Review.
-
Nuclear entry and egress of parvoviruses.Mol Microbiol. 2022 Oct;118(4):295-308. doi: 10.1111/mmi.14974. Epub 2022 Aug 24. Mol Microbiol. 2022. PMID: 35974704 Free PMC article. Review.
Cited by
-
Intranuclear HSV-1 DNA ejection induces major mechanical transformations suggesting mechanoprotection of nucleus integrity.Proc Natl Acad Sci U S A. 2022 Mar 1;119(9):e2114121119. doi: 10.1073/pnas.2114121119. Proc Natl Acad Sci U S A. 2022. PMID: 35197285 Free PMC article.
-
Pressure-driven release of viral genome into a host nucleus is a mechanism leading to herpes infection.Elife. 2019 Aug 8;8:e47212. doi: 10.7554/eLife.47212. Elife. 2019. PMID: 31393262 Free PMC article.
-
Misdelivery at the Nuclear Pore Complex-Stopping a Virus Dead in Its Tracks.Cells. 2015 Jul 28;4(3):277-96. doi: 10.3390/cells4030277. Cells. 2015. PMID: 26226003 Free PMC article. Review.
-
The interferon-inducible GTPase MxB promotes capsid disassembly and genome release of herpesviruses.Elife. 2022 Apr 27;11:e76804. doi: 10.7554/eLife.76804. Elife. 2022. PMID: 35475759 Free PMC article.
-
Pressurized DNA state inside herpes capsids-A novel antiviral target.PLoS Pathog. 2020 Jul 23;16(7):e1008604. doi: 10.1371/journal.ppat.1008604. eCollection 2020 Jul. PLoS Pathog. 2020. PMID: 32702029 Free PMC article.
References
-
- Frenkiel-Krispin D, Maco B, Aebi U, Medalia O. Structural analysis of a metazoan nuclear pore complex reveals a fused concentric ring architecture. J Mol Biol. 2010;395:578–586. - PubMed
-
- Maimon T, Elad N, Dahan I, Medalia O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure. 2012;20:998–1006. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

