A new method to determine in vivo interactomes reveals binding of the Legionella pneumophila effector PieE to multiple rab GTPases
- PMID: 25118235
- PMCID: PMC4145681
- DOI: 10.1128/mBio.01148-14
A new method to determine in vivo interactomes reveals binding of the Legionella pneumophila effector PieE to multiple rab GTPases
Abstract
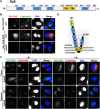
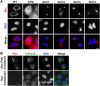
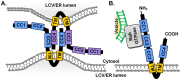
Legionella pneumophila, the causative agent of Legionnaires' disease, uses the Dot/Icm type IV secretion system (T4SS) to translocate more than 300 effectors into host cells, where they subvert host cell signaling. The function and host cell targets of most effectors remain unknown. PieE is a 69-kDa Dot/Icm effector containing three coiled-coil (CC) regions and 2 transmembrane (TM) helices followed by a fourth CC region. Here, we report that PieE dimerized by an interaction between CC3 and CC4. We found that ectopically expressed PieE localized to the endoplasmic reticulum (ER) and induced the formation of organized smooth ER, while following infection PieE localized to the Legionella-containing vacuole (LCV). To identify the physiological targets of PieE during infection, we established a new purification method for which we created an A549 cell line stably expressing the Escherichia coli biotin ligase BirA and infected the cells with L. pneumophila expressing PieE fused to a BirA-specific biotinylation site and a hexahistidine tag. Following tandem Ni(2+) nitrilotriacetic acid (NTA) and streptavidin affinity chromatography, the effector-target complexes were analyzed by mass spectrometry. This revealed interactions of PieE with multiple host cell proteins, including the Rab GTPases 1a, 1b, 2a, 5c, 6a, 7, and 10. Binding of the Rab GTPases, which was validated by yeast two-hybrid binding assays, was mediated by the PieE CC1 and CC2. In summary, using a novel, highly specific strategy to purify effector complexes from infected cells, which is widely applicable to other pathogens, we identified PieE as a multidomain LCV protein with promiscuous Rab GTPase-binding capacity.
Importance: The respiratory pathogen Legionella pneumophila uses the Dot/Icm type IV secretion system to translocate more than 300 effector proteins into host cells. The function of most effectors in infection remains unknown. One of the bottlenecks for their characterization is the identification of target proteins. Frequently used in vitro approaches are not applicable to all effectors and suffer from high rates of false positives or missed interactions, as they are not performed in the context of an infection. Here, we determine key functional domains of the effector PieE and describe a new method to identify host cell targets under physiological infection conditions. Our approach, which is applicable to other pathogens, uncovered the interaction of PieE with several proteins involved in membrane trafficking, in particular Rab GTPases, revealing new details of the Legionella infection strategy and demonstrating the potential of this method to greatly advance our understanding of the molecular basis of infection.
Copyright © 2014 Mousnier et al.
Figures




Similar articles
-
Dot/Icm Effector Translocation by Legionella longbeachae Creates a Replicative Vacuole Similar to That of Legionella pneumophila despite Translocation of Distinct Effector Repertoires.Infect Immun. 2015 Oct;83(10):4081-92. doi: 10.1128/IAI.00461-15. Epub 2015 Jul 27. Infect Immun. 2015. PMID: 26216429 Free PMC article.
-
Legionella Effector AnkX Disrupts Host Cell Endocytic Recycling in a Phosphocholination-Dependent Manner.Front Cell Infect Microbiol. 2017 Sep 8;7:397. doi: 10.3389/fcimb.2017.00397. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28944216 Free PMC article.
-
The Polar Legionella Icm/Dot T4SS Establishes Distinct Contact Sites with the Pathogen Vacuole Membrane.mBio. 2021 Oct 26;12(5):e0218021. doi: 10.1128/mBio.02180-21. Epub 2021 Oct 12. mBio. 2021. PMID: 34634944 Free PMC article.
-
Formation of the Legionella-containing vacuole: phosphoinositide conversion, GTPase modulation and ER dynamics.Int J Med Microbiol. 2018 Jan;308(1):49-57. doi: 10.1016/j.ijmm.2017.08.004. Epub 2017 Aug 16. Int J Med Microbiol. 2018. PMID: 28865995 Review.
-
Manipulation of host vesicular trafficking and innate immune defence by Legionella Dot/Icm effectors.Cell Microbiol. 2011 Dec;13(12):1870-80. doi: 10.1111/j.1462-5822.2011.01710.x. Epub 2011 Nov 10. Cell Microbiol. 2011. PMID: 21981078 Review.
Cited by
-
Dot/Icm Effector Translocation by Legionella longbeachae Creates a Replicative Vacuole Similar to That of Legionella pneumophila despite Translocation of Distinct Effector Repertoires.Infect Immun. 2015 Oct;83(10):4081-92. doi: 10.1128/IAI.00461-15. Epub 2015 Jul 27. Infect Immun. 2015. PMID: 26216429 Free PMC article.
-
Endoplasmic Reticulum Tubule Protein Reticulon 4 Associates with the Legionella pneumophila Vacuole and with Translocated Substrate Ceg9.Infect Immun. 2015 Sep;83(9):3479-89. doi: 10.1128/IAI.00507-15. Epub 2015 Jun 22. Infect Immun. 2015. PMID: 26099580 Free PMC article.
-
The Legionella effector LtpM is a new type of phosphoinositide-activated glucosyltransferase.J Biol Chem. 2019 Feb 22;294(8):2862-2879. doi: 10.1074/jbc.RA118.005952. Epub 2018 Dec 20. J Biol Chem. 2019. PMID: 30573678 Free PMC article.
-
Legionella Effector AnkX Disrupts Host Cell Endocytic Recycling in a Phosphocholination-Dependent Manner.Front Cell Infect Microbiol. 2017 Sep 8;7:397. doi: 10.3389/fcimb.2017.00397. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28944216 Free PMC article.
-
Keeping in Touch with Type-III Secretion System Effectors: Mass Spectrometry-Based Proteomics to Study Effector-Host Protein-Protein Interactions.Int J Mol Sci. 2020 Sep 19;21(18):6891. doi: 10.3390/ijms21186891. Int J Mol Sci. 2020. PMID: 32961832 Free PMC article. Review.
References
-
- Lifshitz Z, Burstein D, Peeri M, Zusman T, Schwartz K, Shuman HA, Pupko T, Segal G. 2013. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc. Natl. Acad. Sci. U. S. A. 110:E707–E715. 10.1073/PNAS1215278110 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

