Genomic alterations in abnormal neutrophils isolated from adult patients with systemic lupus erythematosus
- PMID: 25107306
- PMCID: PMC4262380
- DOI: 10.1186/ar4681
Genomic alterations in abnormal neutrophils isolated from adult patients with systemic lupus erythematosus
Abstract
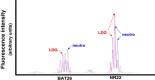
Introduction: Patients with systemic lupus erythematosus (SLE) have an abnormal population of neutrophils, called low-density granulocytes (LDGs), that express the surface markers of mature neutrophils, yet their nuclear morphology resembles an immature cell. Because a similar discrepancy in maturation status is observed in myelodysplasias, and disruption of neutrophil development is frequently associated with genomic alterations, genomic DNA isolated from autologous pairs of LDGs and normal-density neutrophils was compared for genomic changes.
Methods: Alterations in copy number and losses of heterozygosity (LOH) were detected by cytogenetic microarray analysis. Microsatellite instability (MSI) was detected by capillary gel electrophoresis of fluorescently labeled PCR products.
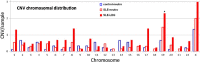
Results: Control neutrophils and normal-density SLE neutrophils had similar levels of copy number variations, while the autologous SLE LDGs had an over twofold greater number of copy number alterations per genome. The additional copy number alterations found in LDGs were prevalent in six of the thirteen SLE patients, and occurred preferentially on chromosome 19, 17, 8, and X. These same SLE patients also displayed an increase in LOH. Several SLE patients had a common LOH on chromosome 5q that includes several cytokine genes and a DNA repair enzyme. In addition, three SLE patients displayed MSI. Two patients displayed MSI in greater than one marker, and one patient had MSI and increased copy number alterations. No correlations between genomic instability and immunosuppressive drugs, disease activity or disease manifestations were apparent.
Conclusions: The increased level of copy number alterations and LOH in the LDG samples relative to autologous normal-density SLE neutrophils suggests somatic alterations that are consistent with DNA strand break repair, while MSI suggests a replication error-prone status. Thus, the LDGs isolated have elevated levels of somatic alterations that are consistent with genetic damage or genomic instability. This suggests that the LDGs in adult SLE patients are derived from cell progenitors that are distinct from the autologous normal-density neutrophils, and may reflect a role for genomic instability in the disease.
Figures



Similar articles
-
Proteomic, biomechanical and functional analyses define neutrophil heterogeneity in systemic lupus erythematosus.Ann Rheum Dis. 2021 Feb;80(2):209-218. doi: 10.1136/annrheumdis-2020-218338. Epub 2020 Sep 28. Ann Rheum Dis. 2021. PMID: 32988843 Free PMC article.
-
Transcriptomic, epigenetic, and functional analyses implicate neutrophil diversity in the pathogenesis of systemic lupus erythematosus.Proc Natl Acad Sci U S A. 2019 Dec 10;116(50):25222-25228. doi: 10.1073/pnas.1908576116. Epub 2019 Nov 21. Proc Natl Acad Sci U S A. 2019. PMID: 31754025 Free PMC article.
-
Genomic Identification of Low-Density Granulocytes and Analysis of Their Role in the Pathogenesis of Systemic Lupus Erythematosus.J Immunol. 2019 Jun 1;202(11):3309-3317. doi: 10.4049/jimmunol.1801512. Epub 2019 Apr 24. J Immunol. 2019. PMID: 31019061
-
Low-density granulocytes in systemic autoimmunity and autoinflammation.Immunol Rev. 2023 Mar;314(1):313-325. doi: 10.1111/imr.13161. Epub 2022 Oct 28. Immunol Rev. 2023. PMID: 36305174 Free PMC article. Review.
-
Neutrophils in the pathogenesis and manifestations of SLE.Nat Rev Rheumatol. 2011 Sep 27;7(12):691-9. doi: 10.1038/nrrheum.2011.132. Nat Rev Rheumatol. 2011. PMID: 21947176 Free PMC article. Review.
Cited by
-
Mycobacterium tuberculosis Infection Induces Low-Density Granulocyte Generation by Promoting Neutrophil Extracellular Trap Formation via ROS Pathway.Front Microbiol. 2019 Jul 11;10:1468. doi: 10.3389/fmicb.2019.01468. eCollection 2019. Front Microbiol. 2019. PMID: 31354639 Free PMC article.
-
Copy number variation in the susceptibility to systemic lupus erythematosus.PLoS One. 2018 Nov 28;13(11):e0206683. doi: 10.1371/journal.pone.0206683. eCollection 2018. PLoS One. 2018. PMID: 30485348 Free PMC article.
-
DNA Damage Response and Immune Defense: Links and Mechanisms.Front Genet. 2016 Aug 9;7:147. doi: 10.3389/fgene.2016.00147. eCollection 2016. Front Genet. 2016. PMID: 27555866 Free PMC article. Review.
-
On the origin of low-density neutrophils.J Leukoc Biol. 2020 May;107(5):809-818. doi: 10.1002/JLB.5HR0120-459R. Epub 2020 Mar 14. J Leukoc Biol. 2020. PMID: 32170882 Free PMC article. Review.
-
Proteomic, biomechanical and functional analyses define neutrophil heterogeneity in systemic lupus erythematosus.Ann Rheum Dis. 2021 Feb;80(2):209-218. doi: 10.1136/annrheumdis-2020-218338. Epub 2020 Sep 28. Ann Rheum Dis. 2021. PMID: 32988843 Free PMC article.
References
-
- Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, Cobb BL, Mirel DB, Marion MC, Williams AH, Divers J, Wang W, Frank SG, Namjou B, Gabriel SB, Lee AT, Gregersen PK, Behrens TW, Taylor KE, Fernando M, Zidovetzki R, Gaffney PM, Edberg JC, Rioux JD, Ojwang JO, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. - DOI - PMC - PubMed
-
- Xu Z, Morel L. Genetics of systemic lupus erythematosus: contributions of mouse models in the era of human genome-wide association studies. Discov Med. 2010;10:71–78. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

