Roles of type 1A topoisomerases in genome maintenance in Escherichia coli
- PMID: 25102178
- PMCID: PMC4125114
- DOI: 10.1371/journal.pgen.1004543
Roles of type 1A topoisomerases in genome maintenance in Escherichia coli
Abstract
In eukaryotes, type 1A topoisomerases (topos) act with RecQ-like helicases to maintain the stability of the genome. Despite having been the first type 1A enzymes to be discovered, much less is known about the involvement of the E. coli topo I (topA) and III (topB) enzymes in genome maintenance. These enzymes are thought to have distinct cellular functions: topo I regulates supercoiling and R-loop formation, and topo III is involved in chromosome segregation. To better characterize their roles in genome maintenance, we have used genetic approaches including suppressor screens, combined with microscopy for the examination of cell morphology and nucleoid shape. We show that topA mutants can suffer from growth-inhibitory and supercoiling-dependent chromosome segregation defects. These problems are corrected by deleting recA or recQ but not by deleting recJ or recO, indicating that the RecF pathway is not involved. Rather, our data suggest that RecQ acts with a type 1A topo on RecA-generated recombination intermediates because: 1-topo III overproduction corrects the defects and 2-recQ deletion and topo IIII overproduction are epistatic to recA deletion. The segregation defects are also linked to over-replication, as they are significantly alleviated by an oriC::aph suppressor mutation which is oriC-competent in topA null but not in isogenic topA+ cells. When both topo I and topo III are missing, excess supercoiling triggers growth inhibition that correlates with the formation of extremely long filaments fully packed with unsegregated and diffuse DNA. These phenotypes are likely related to replication from R-loops as they are corrected by overproducing RNase HI or by genetic suppressors of double topA rnhA mutants affecting constitutive stable DNA replication, dnaT::aph and rne::aph, which initiates from R-loops. Thus, bacterial type 1A topos maintain the stability of the genome (i) by preventing over-replication originating from oriC (topo I alone) and R-loops and (ii) by acting with RecQ.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

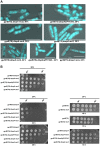

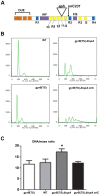


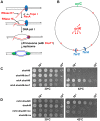

Similar articles
-
Constitutive stable DNA replication in Escherichia coli cells lacking type 1A topoisomerase activity.DNA Repair (Amst). 2015 Nov;35:37-47. doi: 10.1016/j.dnarep.2015.08.004. Epub 2015 Sep 26. DNA Repair (Amst). 2015. PMID: 26444226
-
Mutations reducing replication from R-loops suppress the defects of growth, chromosome segregation and DNA supercoiling in cells lacking topoisomerase I and RNase HI activity.DNA Repair (Amst). 2016 Apr;40:1-17. doi: 10.1016/j.dnarep.2016.02.001. Epub 2016 Feb 27. DNA Repair (Amst). 2016. PMID: 26947024
-
Topoisomerases I and III inhibit R-loop formation to prevent unregulated replication in the chromosomal Ter region of Escherichia coli.PLoS Genet. 2018 Sep 17;14(9):e1007668. doi: 10.1371/journal.pgen.1007668. eCollection 2018 Sep. PLoS Genet. 2018. PMID: 30222737 Free PMC article.
-
Supercoiling, R-loops, Replication and the Functions of Bacterial Type 1A Topoisomerases.Genes (Basel). 2020 Feb 27;11(3):249. doi: 10.3390/genes11030249. Genes (Basel). 2020. PMID: 32120891 Free PMC article. Review.
-
R-loop-dependent replication and genomic instability in bacteria.DNA Repair (Amst). 2019 Dec;84:102693. doi: 10.1016/j.dnarep.2019.102693. Epub 2019 Aug 21. DNA Repair (Amst). 2019. PMID: 31471263 Review.
Cited by
-
A highly processive actinobacterial topoisomerase I - thoughts on Streptomyces' demand for an enzyme with a unique C-terminal domain.Microbiology (Reading). 2020 Feb;166(2):120-128. doi: 10.1099/mic.0.000841. Epub 2019 Aug 7. Microbiology (Reading). 2020. PMID: 31390324 Free PMC article. Review.
-
Non-Canonical Replication Initiation: You're Fired!Genes (Basel). 2017 Jan 27;8(2):54. doi: 10.3390/genes8020054. Genes (Basel). 2017. PMID: 28134821 Free PMC article. Review.
-
Characterization of a pathway of genomic instability induced by R-loops and its regulation by topoisomerases in E. coli.PLoS Genet. 2023 May 4;19(5):e1010754. doi: 10.1371/journal.pgen.1010754. eCollection 2023 May. PLoS Genet. 2023. PMID: 37141391 Free PMC article.
-
Covalent Complex of DNA and Bacterial Topoisomerase: Implications in Antibacterial Drug Development.ChemMedChem. 2020 Apr 3;15(7):623-631. doi: 10.1002/cmdc.201900721. Epub 2020 Mar 18. ChemMedChem. 2020. PMID: 32043806 Free PMC article.
-
TopA, the Sulfolobus solfataricus topoisomerase III, is a decatenase.Nucleic Acids Res. 2018 Jan 25;46(2):861-872. doi: 10.1093/nar/gkx1247. Nucleic Acids Res. 2018. PMID: 29253195 Free PMC article.
References
-
- Chen SH, Chan NL, Hsieh TS (2013) New mechanistic and functional insights into DNA topoisomerases. Annu Rev Biochem 82: 139–170. - PubMed
-
- Champoux JJ (2001) DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem 70: 369–413. - PubMed
-
- Wang JC (1971) Interaction between DNA and an Escherichia coli protein omega. J Mol Biol 55: 523–533. - PubMed
-
- Kirkegaard K, Wang JC (1985) Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single-stranded loop. J Mol Biol 185: 625–637. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

