Antifungal drug resistance evoked via RNAi-dependent epimutations
- PMID: 25079329
- PMCID: PMC4177005
- DOI: 10.1038/nature13575
Antifungal drug resistance evoked via RNAi-dependent epimutations
Abstract
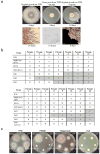
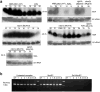
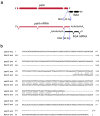
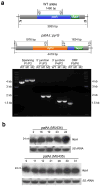
Microorganisms evolve via a range of mechanisms that may include or involve sexual/parasexual reproduction, mutators, aneuploidy, Hsp90 and even prions. Mechanisms that may seem detrimental can be repurposed to generate diversity. Here we show that the human fungal pathogen Mucor circinelloides develops spontaneous resistance to the antifungal drug FK506 (tacrolimus) via two distinct mechanisms. One involves Mendelian mutations that confer stable drug resistance; the other occurs via an epigenetic RNA interference (RNAi)-mediated pathway resulting in unstable drug resistance. The peptidylprolyl isomerase FKBP12 interacts with FK506 forming a complex that inhibits the protein phosphatase calcineurin. Calcineurin inhibition by FK506 blocks M. circinelloides transition to hyphae and enforces yeast growth. Mutations in the fkbA gene encoding FKBP12 or the calcineurin cnbR or cnaA genes confer FK506 resistance and restore hyphal growth. In parallel, RNAi is spontaneously triggered to silence the fkbA gene, giving rise to drug-resistant epimutants. FK506-resistant epimutants readily reverted to the drug-sensitive wild-type phenotype when grown without exposure to the drug. The establishment of these epimutants is accompanied by generation of abundant fkbA small RNAs and requires the RNAi pathway as well as other factors that constrain or reverse the epimutant state. Silencing involves the generation of a double-stranded RNA trigger intermediate using the fkbA mature mRNA as a template to produce antisense fkbA RNA. This study uncovers a novel epigenetic RNAi-based epimutation mechanism controlling phenotypic plasticity, with possible implications for antimicrobial drug resistance and RNAi-regulatory mechanisms in fungi and other eukaryotes.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
Drug-Resistant Epimutants Exhibit Organ-Specific Stability and Induction during Murine Infections Caused by the Human Fungal Pathogen Mucor circinelloides.mBio. 2019 Nov 5;10(6):e02579-19. doi: 10.1128/mBio.02579-19. mBio. 2019. PMID: 31690679 Free PMC article.
-
Broad antifungal resistance mediated by RNAi-dependent epimutation in the basal human fungal pathogen Mucor circinelloides.PLoS Genet. 2019 Feb 11;15(2):e1007957. doi: 10.1371/journal.pgen.1007957. eCollection 2019 Feb. PLoS Genet. 2019. PMID: 30742617 Free PMC article.
-
A Novel Resistance Pathway for Calcineurin Inhibitors in the Human-Pathogenic Mucorales Mucor circinelloides.mBio. 2020 Jan 28;11(1):e02949-19. doi: 10.1128/mBio.02949-19. mBio. 2020. PMID: 31992620 Free PMC article.
-
Calcineurin in fungal virulence and drug resistance: Prospects for harnessing targeted inhibition of calcineurin for an antifungal therapeutic approach.Virulence. 2017 Feb 17;8(2):186-197. doi: 10.1080/21505594.2016.1201250. Epub 2016 Jun 20. Virulence. 2017. PMID: 27325145 Free PMC article. Review.
-
Distinct RNAi Pathways in the Regulation of Physiology and Development in the Fungus Mucor circinelloides.Adv Genet. 2015;91:55-102. doi: 10.1016/bs.adgen.2015.07.002. Epub 2015 Aug 7. Adv Genet. 2015. PMID: 26410030 Review.
Cited by
-
Calcineurin orchestrates dimorphic transitions, antifungal drug responses and host-pathogen interactions of the pathogenic mucoralean fungus Mucor circinelloides.Mol Microbiol. 2015 Sep;97(5):844-65. doi: 10.1111/mmi.13071. Epub 2015 Jun 17. Mol Microbiol. 2015. PMID: 26010100 Free PMC article.
-
The RNAi machinery controls distinct responses to environmental signals in the basal fungus Mucor circinelloides.BMC Genomics. 2015 Mar 25;16(1):237. doi: 10.1186/s12864-015-1443-2. BMC Genomics. 2015. PMID: 25880254 Free PMC article.
-
Heterochromatin and RNAi act independently to ensure genome stability in Mucorales human fungal pathogens.Proc Natl Acad Sci U S A. 2023 Feb 14;120(7):e2220475120. doi: 10.1073/pnas.2220475120. Epub 2023 Feb 6. Proc Natl Acad Sci U S A. 2023. PMID: 36745785 Free PMC article.
-
Recent Advances and Future Directions in the Understanding of Mucormycosis.Front Cell Infect Microbiol. 2022 Feb 24;12:850581. doi: 10.3389/fcimb.2022.850581. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35281441 Free PMC article. Review.
-
Biologia futura: combinatorial stress responses in fungi.Biol Futur. 2022 Jun;73(2):207-217. doi: 10.1007/s42977-022-00121-8. Epub 2022 Jun 15. Biol Futur. 2022. PMID: 35704178 Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

