Structural insights into the catalytic mechanism of Synechocystis magnesium protoporphyrin IX O-methyltransferase (ChlM)
- PMID: 25077963
- PMCID: PMC4162172
- DOI: 10.1074/jbc.M114.584920
Structural insights into the catalytic mechanism of Synechocystis magnesium protoporphyrin IX O-methyltransferase (ChlM)
Abstract
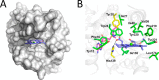
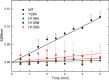
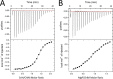
Magnesium protoporphyrin IX O-methyltransferase (ChlM) catalyzes transfer of the methyl group from S-adenosylmethionine to the carboxyl group of the C13 propionate side chain of magnesium protoporphyrin IX. This reaction is the second committed step in chlorophyll biosynthesis from protoporphyrin IX. Here we report the crystal structures of ChlM from the cyanobacterium Synechocystis sp. PCC 6803 in complex with S-adenosylmethionine and S-adenosylhomocysteine at resolutions of 1.6 and 1.7 Å, respectively. The structures illustrate the molecular basis for cofactor and substrate binding and suggest that conformational changes of the two "arm" regions may modulate binding and release of substrates/products to and from the active site. Tyr-28 and His-139 were identified to play essential roles for methyl transfer reaction but are not indispensable for cofactor/substrate binding. Based on these structural and functional findings, a catalytic model is proposed.
Keywords: Chlorophyll; Chloroplast; Enzyme Mechanism; Enzyme Structure; Photosynthesis; Protein Structure.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


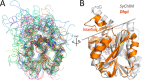
Similar articles
-
An enzyme-coupled continuous spectrophotometric assay for magnesium protoporphyrin IX methyltransferases.Anal Biochem. 2009 Nov 15;394(2):223-8. doi: 10.1016/j.ab.2009.07.036. Epub 2009 Jul 29. Anal Biochem. 2009. PMID: 19646414
-
Introduction of a new branchpoint in tetrapyrrole biosynthesis in Escherichia coli by co-expression of genes encoding the chlorophyll-specific enzymes magnesium chelatase and magnesium protoporphyrin methyltransferase.FEBS Lett. 1999 Jul 23;455(3):349-54. doi: 10.1016/s0014-5793(99)00909-6. FEBS Lett. 1999. PMID: 10437802
-
Knock-out of the magnesium protoporphyrin IX methyltransferase gene in Arabidopsis. Effects on chloroplast development and on chloroplast-to-nucleus signaling.J Biol Chem. 2007 Jan 26;282(4):2297-304. doi: 10.1074/jbc.M610286200. Epub 2006 Nov 29. J Biol Chem. 2007. PMID: 17135235 Free PMC article.
-
Methyltransferase Inhibitors: Competing with, or Exploiting the Bound Cofactor.Molecules. 2019 Dec 8;24(24):4492. doi: 10.3390/molecules24244492. Molecules. 2019. PMID: 31817960 Free PMC article. Review.
-
Structure, function and physiological role of glycine N-methyltransferase.Int J Biochem Cell Biol. 1998 Jan;30(1):13-26. doi: 10.1016/s1357-2725(97)00105-2. Int J Biochem Cell Biol. 1998. PMID: 9597750 Review.
Cited by
-
Mutagenesis selection and large-scale cultivation of non-green Chlamydomonas reinhardtii for food applications.Front Nutr. 2024 Sep 25;11:1456230. doi: 10.3389/fnut.2024.1456230. eCollection 2024. Front Nutr. 2024. PMID: 39385786 Free PMC article.
-
A Thylakoid Membrane Protein Functions Synergistically with GUN5 in Chlorophyll Biosynthesis.Plant Commun. 2020 Jul 3;1(5):100094. doi: 10.1016/j.xplc.2020.100094. eCollection 2020 Sep 14. Plant Commun. 2020. PMID: 33367259 Free PMC article.
-
The terminal enzymes of (bacterio)chlorophyll biosynthesis.R Soc Open Sci. 2022 May 4;9(5):211903. doi: 10.1098/rsos.211903. eCollection 2022 May. R Soc Open Sci. 2022. PMID: 35573041 Free PMC article. Review.
-
Biosynthesis of the modified tetrapyrroles-the pigments of life.J Biol Chem. 2020 May 15;295(20):6888-6925. doi: 10.1074/jbc.REV120.006194. Epub 2020 Apr 2. J Biol Chem. 2020. PMID: 32241908 Free PMC article. Review.
-
Molecular mechanism of S-adenosylmethionine sensing by SAMTOR in mTORC1 signaling.Sci Adv. 2022 Jul;8(26):eabn3868. doi: 10.1126/sciadv.abn3868. Epub 2022 Jul 1. Sci Adv. 2022. PMID: 35776786 Free PMC article.
References
-
- Chew A. G., Bryant D. A. (2007) Chlorophyll biosynthesis in bacteria: the origins of structural and functional diversity. Annu. Rev. Microbiol. 61, 113–129 - PubMed
-
- Tanaka R., Tanaka A. (2007) Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58, 321–346 - PubMed
-
- Mochizuki N., Tanaka R., Grimm B., Masuda T., Moulin M., Smith A. G., Tanaka A., Terry M. J. (2010) The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Sci. 15, 488–498 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

