Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas
- PMID: 25056374
- PMCID: PMC4305034
- DOI: 10.1158/1078-0432.CCR-13-3310
Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas
Abstract
Purpose: The genetic differences between human papilloma virus (HPV)-positive and -negative head and neck squamous cell carcinomas (HNSCC) remain largely unknown. To identify differential biology and novel therapeutic targets for both entities, we determined mutations and copy-number aberrations in a large cohort of locoregionally advanced HNSCC.
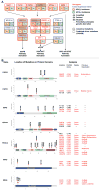
Experimental design: We performed massively parallel sequencing of 617 cancer-associated genes in 120 matched tumor/normal samples (42.5% HPV-positive). Mutations and copy-number aberrations were determined and results validated with a secondary method.
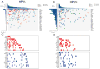
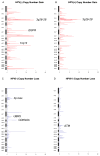
Results: The overall mutational burden in HPV-negative and HPV-positive HNSCC was similar with an average of 15.2 versus 14.4 somatic exonic mutations in the targeted cancer-associated genes. HPV-negative tumors showed a mutational spectrum concordant with published lung squamous cell carcinoma analyses with enrichment for mutations in TP53, CDKN2A, MLL2, CUL3, NSD1, PIK3CA, and NOTCH genes. HPV-positive tumors showed unique mutations in DDX3X, FGFR2/3 and aberrations in PIK3CA, KRAS, MLL2/3, and NOTCH1 were enriched in HPV-positive tumors. Currently targetable genomic alterations were identified in FGFR1, DDR2, EGFR, FGFR2/3, EPHA2, and PIK3CA. EGFR, CCND1, and FGFR1 amplifications occurred in HPV-negative tumors, whereas 17.6% of HPV-positive tumors harbored mutations in fibroblast growth factor receptor genes (FGFR2/3), including six recurrent FGFR3 S249C mutations. HPV-positive tumors showed a 5.8% incidence of KRAS mutations, and DNA-repair gene aberrations, including 7.8% BRCA1/2 mutations, were identified.
Conclusions: The mutational makeup of HPV-positive and HPV-negative HNSCC differs significantly, including targetable genes. HNSCC harbors multiple therapeutically important genetic aberrations, including frequent aberrations in the FGFR and PI3K pathway genes. See related commentary by Krigsfeld and Chung, p. 495.
©2014 American Association for Cancer Research.
Conflict of interest statement
The authors report no conflict of interest.
Figures
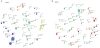
Comment in
-
Novel targets in head and neck cancer: should we be optimistic?Clin Cancer Res. 2015 Feb 1;21(3):495-7. doi: 10.1158/1078-0432.CCR-14-1776. Epub 2014 Oct 9. Clin Cancer Res. 2015. PMID: 25301846 Free PMC article.
Similar articles
-
Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing.Ann Oncol. 2015 Jun;26(6):1216-1223. doi: 10.1093/annonc/mdv109. Epub 2015 Feb 23. Ann Oncol. 2015. PMID: 25712460 Free PMC article.
-
Comprehensive genomic profiling of head and neck squamous cell carcinoma reveals FGFR1 amplifications and tumour genomic alterations burden as prognostic biomarkers of survival.Eur J Cancer. 2018 Mar;91:47-55. doi: 10.1016/j.ejca.2017.12.016. Epub 2018 Jan 11. Eur J Cancer. 2018. PMID: 29331751
-
Distinct pattern of TP53 mutations in human immunodeficiency virus-related head and neck squamous cell carcinoma.Cancer. 2018 Jan 1;124(1):84-94. doi: 10.1002/cncr.31063. Epub 2017 Oct 20. Cancer. 2018. PMID: 29053175 Free PMC article.
-
[Genomic characterization of head and neck squamous cell carcinoma: Impact and challenges for therapeutic management].Bull Cancer. 2018 Sep;105(9):820-829. doi: 10.1016/j.bulcan.2018.05.011. Epub 2018 Jun 22. Bull Cancer. 2018. PMID: 29937334 Review. French.
-
Integrating genomics in head and neck cancer treatment: Promises and pitfalls.Crit Rev Oncol Hematol. 2015 Sep;95(3):397-406. doi: 10.1016/j.critrevonc.2015.03.005. Epub 2015 Apr 18. Crit Rev Oncol Hematol. 2015. PMID: 25979769 Review.
Cited by
-
Crosstalk between CAFs and tumour cells in head and neck cancer.Cell Death Discov. 2024 Jun 26;10(1):303. doi: 10.1038/s41420-024-02053-9. Cell Death Discov. 2024. PMID: 38926351 Free PMC article. Review.
-
Ex vivo radiation sensitivity assessment for individual head and neck cancer patients using deep learning-based automated nuclei and DNA damage foci detection.Clin Transl Radiat Oncol. 2024 Jan 30;45:100735. doi: 10.1016/j.ctro.2024.100735. eCollection 2024 Mar. Clin Transl Radiat Oncol. 2024. PMID: 38380115 Free PMC article.
-
Human papillomavirus oncoprotein E6 upregulates c-Met through p53 downregulation.Eur J Cancer. 2016 Sep;65:21-32. doi: 10.1016/j.ejca.2016.06.006. Epub 2016 Jul 22. Eur J Cancer. 2016. PMID: 27451021 Free PMC article.
-
Notch Signaling and Human Papillomavirus-Associated Oral Tumorigenesis.Adv Exp Med Biol. 2021;1287:105-122. doi: 10.1007/978-3-030-55031-8_8. Adv Exp Med Biol. 2021. PMID: 33034029 Free PMC article. Review.
-
Improved outcomes in PI3K-pathway-altered metastatic HPV oropharyngeal cancer.JCI Insight. 2018 Sep 6;3(17):e122799. doi: 10.1172/jci.insight.122799. eCollection 2018 Sep 6. JCI Insight. 2018. PMID: 30185662 Free PMC article.
References
-
- Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. - PubMed
-
- Vokes EE, Weichselbaum RR, Lippman SM, Hong WK. Head and neck cancer. N Engl J Med. 1993;328:184–194. - PubMed
-
- Hayes David N, Grandis Jennifer R, El-Naggar Adel K. The Cancer Genome Atlas: Integrated analysis of genome alterations in squamous cell carcinoma of the head and neck. J Clin Oncol. 2013;31(suppl):abstr 6009.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

