dREAM co-operates with insulator-binding proteins and regulates expression at divergently paired genes
- PMID: 25053843
- PMCID: PMC4132727
- DOI: 10.1093/nar/gku609
dREAM co-operates with insulator-binding proteins and regulates expression at divergently paired genes
Abstract
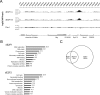
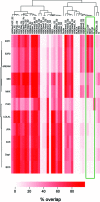
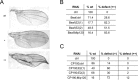
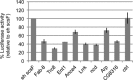
dREAM complexes represent the predominant form of E2F/RBF repressor complexes in Drosophila. dREAM associates with thousands of sites in the fly genome but its mechanism of action is unknown. To understand the genomic context in which dREAM acts we examined the distribution and localization of Drosophila E2F and dREAM proteins. Here we report a striking and unexpected overlap between dE2F2/dREAM sites and binding sites for the insulator-binding proteins CP190 and Beaf-32. Genetic assays show that these components functionally co-operate and chromatin immunoprecipitation experiments on mutant animals demonstrate that dE2F2 is important for association of CP190 with chromatin. dE2F2/dREAM binding sites are enriched at divergently transcribed genes, and the majority of genes upregulated by dE2F2 depletion represent the repressed half of a differentially expressed, divergently transcribed pair of genes. Analysis of mutant animals confirms that dREAM and CP190 are similarly required for transcriptional integrity at these gene pairs and suggest that dREAM functions in concert with CP190 to establish boundaries between repressed/activated genes. Consistent with the idea that dREAM co-operates with insulator-binding proteins, genomic regions bound by dREAM possess enhancer-blocking activity that depends on multiple dREAM components. These findings suggest that dREAM functions in the organization of transcriptional domains.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
A functional insulator screen identifies NURF and dREAM components to be required for enhancer-blocking.PLoS One. 2014 Sep 23;9(9):e107765. doi: 10.1371/journal.pone.0107765. eCollection 2014. PLoS One. 2014. PMID: 25247414 Free PMC article.
-
Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin.Genome Res. 2015 Jan;25(1):89-99. doi: 10.1101/gr.174169.114. Epub 2014 Oct 23. Genome Res. 2015. PMID: 25342723 Free PMC article.
-
Genome-wide localization of exosome components to active promoters and chromatin insulators in Drosophila.Nucleic Acids Res. 2013 Mar 1;41(5):2963-80. doi: 10.1093/nar/gkt037. Epub 2013 Jan 28. Nucleic Acids Res. 2013. PMID: 23358822 Free PMC article.
-
Functional sub-division of the Drosophila genome via chromatin looping: the emerging importance of CP190.Nucleus. 2013 Mar-Apr;4(2):115-22. doi: 10.4161/nucl.23389. Epub 2013 Jan 18. Nucleus. 2013. PMID: 23333867 Free PMC article. Review.
-
DREAM a little dREAM of DRM: Model organisms and conservation of DREAM-like complexes: Model organisms uncover the mechanisms of DREAM-mediated transcription regulation.Bioessays. 2024 Feb;46(2):e2300125. doi: 10.1002/bies.202300125. Epub 2023 Dec 7. Bioessays. 2024. PMID: 38059789 Review.
Cited by
-
Structure and function of MuvB complexes.Oncogene. 2022 May;41(21):2909-2919. doi: 10.1038/s41388-022-02321-x. Epub 2022 Apr 26. Oncogene. 2022. PMID: 35468940 Free PMC article. Review.
-
Mechanism and functional role of the interaction between CP190 and the architectural protein Pita in Drosophila melanogaster.Epigenetics Chromatin. 2021 Mar 22;14(1):16. doi: 10.1186/s13072-021-00391-x. Epigenetics Chromatin. 2021. PMID: 33752739 Free PMC article.
-
Binding profiles for 954 Drosophila and C. elegans transcription factors reveal tissue specific regulatory relationships.bioRxiv [Preprint]. 2024 Jan 20:2024.01.18.576242. doi: 10.1101/2024.01.18.576242. bioRxiv. 2024. Update in: Genome Res. 2024 Dec 23;34(12):2319-2334. doi: 10.1101/gr.279037.124. PMID: 38293065 Free PMC article. Updated. Preprint.
-
Newly Characterized Region of CP190 Associates with Microtubules and Mediates Proper Spindle Morphology in Drosophila Stem Cells.PLoS One. 2015 Dec 9;10(12):e0144174. doi: 10.1371/journal.pone.0144174. eCollection 2015. PLoS One. 2015. PMID: 26649574 Free PMC article.
-
Genome-Wide Analysis of Drosophila RBf2 Protein Highlights the Diversity of RB Family Targets and Possible Role in Regulation of Ribosome Biosynthesis.G3 (Bethesda). 2015 May 20;5(7):1503-15. doi: 10.1534/g3.115.019166. G3 (Bethesda). 2015. PMID: 25999584 Free PMC article.
References
-
- Cooper K. Rb, whi it's not just for metazoans anymore. Oncogene. 2006;25:5228–5232. - PubMed
-
- Gutzat R., Borghi L., Gruissem W. Emerging roles of RETINOBLASTOMA-RELATED proteins in evolution and plant development. Trends Plant Sci. 2012;17:139–148. - PubMed
-
- van den Heuvel S., Dyson N.J. Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 2008;9:713–724. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

