Molecular mechanism of allosteric modulation at GPCRs: insight from a binding kinetics study at the human A1 adenosine receptor
- PMID: 25040887
- PMCID: PMC4294041
- DOI: 10.1111/bph.12836
Molecular mechanism of allosteric modulation at GPCRs: insight from a binding kinetics study at the human A1 adenosine receptor
Abstract
Background and purpose: Many GPCRs can be allosterically modulated by small-molecule ligands. This modulation is best understood in terms of the kinetics of the ligand-receptor interaction. However, many current kinetic assays require at least the (radio)labelling of the orthosteric ligand, which is impractical for studying a range of ligands. Here, we describe the application of a so-called competition association assay at the adenosine A1 receptor for this purpose.
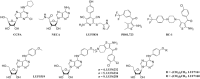
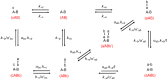
Experimental approach: We used a competition association assay to examine the binding kinetics of several unlabelled orthosteric agonists of the A1 receptor in the absence or presence of two allosteric modulators. We also tested three bitopic ligands, in which an orthosteric and an allosteric pharmacophore were covalently linked with different spacer lengths. The relevance of the competition association assay for the binding kinetics of the bitopic ligands was also explored by analysing simulated data.
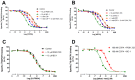
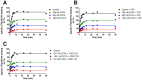
Key results: The binding kinetics of an unlabelled orthosteric ligand were affected by the addition of an allosteric modulator and such effects were probe- and concentration-dependent. Covalently linking the orthosteric and allosteric pharmacophores into one bitopic molecule had a substantial effect on the overall on- or off-rate.
Conclusion and implications: The competition association assay is a useful tool for exploring the allosteric modulation of the human adenosine A1 receptor. This assay may have general applicability to study allosteric modulation at other GPCRs as well.
© 2014 The British Pharmacological Society.
Figures


Similar articles
-
Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Enabling Systemic Identification and Functionality Profiling for Cdc42 Homeostatic Modulators.bioRxiv [Preprint]. 2024 Jan 8:2024.01.05.574351. doi: 10.1101/2024.01.05.574351. bioRxiv. 2024. Update in: Commun Chem. 2024 Nov 19;7(1):271. doi: 10.1038/s42004-024-01352-7. PMID: 38260445 Free PMC article. Updated. Preprint.
-
Interventions to reduce harm from continued tobacco use.Cochrane Database Syst Rev. 2016 Oct 13;10(10):CD005231. doi: 10.1002/14651858.CD005231.pub3. Cochrane Database Syst Rev. 2016. PMID: 27734465 Free PMC article. Review.
Cited by
-
Species dependence of A3 adenosine receptor pharmacology and function.Purinergic Signal. 2023 Sep;19(3):523-550. doi: 10.1007/s11302-022-09910-1. Epub 2022 Dec 20. Purinergic Signal. 2023. PMID: 36538251 Free PMC article. Review.
-
Analysis of equilibrium binding of an orthosteric tracer and two allosteric modulators.PLoS One. 2019 Mar 27;14(3):e0214255. doi: 10.1371/journal.pone.0214255. eCollection 2019. PLoS One. 2019. PMID: 30917186 Free PMC article.
-
Nanobody-Mediated Dualsteric Engagement of the Angiotensin Receptor Broadens Biased Ligand Pharmacology.Mol Pharmacol. 2024 Feb 15;105(3):260-271. doi: 10.1124/molpharm.123.000797. Mol Pharmacol. 2024. PMID: 38164609 Free PMC article.
-
Mass spectrometry-based ligand binding assays on adenosine A1 and A2A receptors.Purinergic Signal. 2015 Dec;11(4):581-94. doi: 10.1007/s11302-015-9477-0. Epub 2015 Oct 19. Purinergic Signal. 2015. PMID: 26482925 Free PMC article.
-
Hit-to-lead and lead optimization binding free energy calculations for G protein-coupled receptors.Interface Focus. 2020 Dec 6;10(6):20190128. doi: 10.1098/rsfs.2019.0128. Epub 2020 Oct 16. Interface Focus. 2020. PMID: 33178414 Free PMC article.
References
-
- Beukers MW, Chang LC, von Frijtag Drabbe Kunzel JK, Mulder-Krieger T, Spanjersberg RF, Brussee J, et al. New, non-adenosine, high-potency agonists for the human adenosine A2B receptor with an improved selectivity profile compared to the reference agonist N-ethylcarboxamidoadenosine. J Med Chem. 2004;47:3707–3709. - PubMed
-
- Bhattacharya S, Linden J. The allosteric enhancer, PD 81,723, stabilizes human A1 adenosine receptor coupling to G proteins. Biochim Biophys Acta. 1995;1265:15–21. - PubMed
-
- Bruns RF, Fergus JH. Allosteric enhancement of adenosine A1 receptor binding and function by 2-amino-3-benzoylthiophenes. Mol Pharmacol. 1990;38:939–949. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

