CREB SUMOylation by the E3 ligase PIAS1 enhances spatial memory
- PMID: 25031400
- PMCID: PMC6608321
- DOI: 10.1523/JNEUROSCI.4302-13.2014
CREB SUMOylation by the E3 ligase PIAS1 enhances spatial memory
Abstract
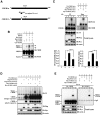
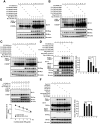
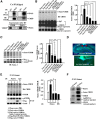
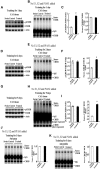
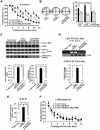
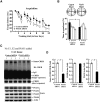
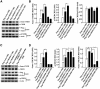
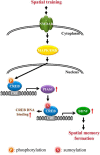
cAMP-responsive element binding protein (CREB) phosphorylation and signaling plays an important role in long-term memory formation, but other posttranslational modifications of CREB are less known. Here, we found that CREB1Δ, the short isoform of CREB, could be sumoylated by the small ubiquitin-like modifier (SUMO) E3 ligase protein inhibitor of activated STAT1 (PIAS1) at Lys271 and Lys290 and PIAS1 SUMOylation of CREB1Δ increased the expression level of CREB1Δ. CREB1Δ could also be sumoylated by other PIAS family proteins, but not by the E3 ligases RanBP2 and Pc2 or by the E2 ligase Ubc9. Furthermore, water maze training increased the level of endogenous CREB SUMOylation in rat CA1 neurons determined by in vitro SUMOylation assay, but this effect was not observed in other brain areas. Moreover, transduction of Lenti-CREBWT to rat CA1 area facilitated, whereas transduction of Lenti-CREB double sumo-mutant (CREBK271RK290R) impaired, spatial learning and memory performance. Transduction of Lenti-CREBWT-SUMO1 fusion vector to rat CA1 area showed a more significant effect in enhancing spatial learning and memory and CREB SUMOylation. Lenti-CREBWT transduction increased, whereas Lenti-CREBK271RK290R transduction decreased, CREB DNA binding to the brain-derived neurotrophic factor (bdnf) promoter and decreased bdnf mRNA expression. Knock-down of PIAS1 expression in CA1 area by PIAS1 siRNA transfection impaired spatial learning and memory and decreased endogenous CREB SUMOylation. In addition, CREB SUMOylation was CREB phosphorylation dependent and lasted longer. Therefore, CREB phosphorylation may be responsible for signal transduction during the early phase of long-term memory formation, whereas CREB SUMOylation sustains long-term memory.
Keywords: CREB; PIAS1; SUMOylation; phosphorylation; spatial learning and memory.
Copyright © 2014 the authors 0270-6474/14/349574-16$15.00/0.
Figures

Similar articles
-
Protein inhibitor of activated STAT1 Ser503 phosphorylation-mediated Elk-1 SUMOylation promotes neuronal survival in APP/PS1 mice.Br J Pharmacol. 2019 Jun;176(11):1793-1810. doi: 10.1111/bph.14656. Epub 2019 Apr 24. Br J Pharmacol. 2019. PMID: 30849179 Free PMC article.
-
Novel role and mechanism of protein inhibitor of activated STAT1 in spatial learning.EMBO J. 2011 Jan 5;30(1):205-20. doi: 10.1038/emboj.2010.290. Epub 2010 Nov 19. EMBO J. 2011. PMID: 21102409 Free PMC article.
-
Phosphorylation of protein inhibitor of activated STAT1 (PIAS1) by MAPK-activated protein kinase-2 inhibits endothelial inflammation via increasing both PIAS1 transrepression and SUMO E3 ligase activity.Arterioscler Thromb Vasc Biol. 2013 Feb;33(2):321-9. doi: 10.1161/ATVBAHA.112.300619. Epub 2012 Nov 29. Arterioscler Thromb Vasc Biol. 2013. PMID: 23202365 Free PMC article.
-
Regulation of CREB Functions by Phosphorylation and Sumoylation in Nervous and Visual Systems.Curr Mol Med. 2017;16(10):885-892. doi: 10.2174/1566524016666161223110106. Curr Mol Med. 2017. PMID: 28017136 Review.
-
PIAS family in cancer: from basic mechanisms to clinical applications.Front Oncol. 2024 Mar 25;14:1376633. doi: 10.3389/fonc.2024.1376633. eCollection 2024. Front Oncol. 2024. PMID: 38590645 Free PMC article. Review.
Cited by
-
Hypoxia-mediated alterations and their role in the HER-2/neuregulated CREB status and localization.Oncotarget. 2016 Aug 9;7(32):52061-52084. doi: 10.18632/oncotarget.10474. Oncotarget. 2016. PMID: 27409833 Free PMC article.
-
Protein inhibitor of activated STAT1 (PIAS1) alleviates cerebral infarction and inflammation after cerebral ischemia in rats.Heliyon. 2024 Jan 14;10(7):e24743. doi: 10.1016/j.heliyon.2024.e24743. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38617924 Free PMC article.
-
Super-resolution study of PIAS SUMO E3-ligases in hippocampal and cortical neurons.Eur J Histochem. 2021 Aug 11;65(s1):3241. doi: 10.4081/ejh.2021.3241. Eur J Histochem. 2021. PMID: 34459572 Free PMC article.
-
Novel insights into the impact of the SUMOylation pathway in hematological malignancies (Review).Int J Oncol. 2021 Sep;59(3):73. doi: 10.3892/ijo.2021.5253. Epub 2021 Aug 9. Int J Oncol. 2021. PMID: 34368858 Free PMC article. Review.
-
Protein inhibitor of activated STAT1 Ser503 phosphorylation-mediated Elk-1 SUMOylation promotes neuronal survival in APP/PS1 mice.Br J Pharmacol. 2019 Jun;176(11):1793-1810. doi: 10.1111/bph.14656. Epub 2019 Apr 24. Br J Pharmacol. 2019. PMID: 30849179 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
