Dynamics of linker residues modulate the nucleic acid binding properties of the HIV-1 nucleocapsid protein zinc fingers
- PMID: 25029439
- PMCID: PMC4100767
- DOI: 10.1371/journal.pone.0102150
Dynamics of linker residues modulate the nucleic acid binding properties of the HIV-1 nucleocapsid protein zinc fingers
Abstract
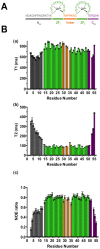
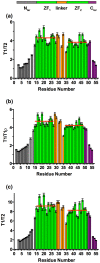
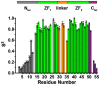
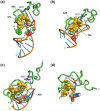
The HIV-1 nucleocapsid protein (NC) is a small basic protein containing two zinc fingers (ZF) separated by a short linker. It is involved in several steps of the replication cycle and acts as a nucleic acid chaperone protein in facilitating nucleic acid strand transfers occurring during reverse transcription. Recent analysis of three-dimensional structures of NC-nucleic acids complexes established a new property: the unpaired guanines targeted by NC are more often inserted in the C-terminal zinc finger (ZF2) than in the N-terminal zinc finger (ZF1). Although previous NMR dynamic studies were performed with NC, the dynamic behavior of the linker residues connecting the two ZF domains remains unclear. This prompted us to investigate the dynamic behavior of the linker residues. Here, we collected 15N NMR relaxation data and used for the first time data at several fields to probe the protein dynamics. The analysis at two fields allows us to detect a slow motion occurring between the two domains around a hinge located in the linker at the G35 position. However, the amplitude of motion appears limited in our conditions. In addition, we showed that the neighboring linker residues R29, A30, P31, R32, K33 displayed restricted motion and numerous contacts with residues of ZF1. Our results are fully consistent with a model in which the ZF1-linker contacts prevent the ZF1 domain to interact with unpaired guanines, whereas the ZF2 domain is more accessible and competent to interact with unpaired guanines. In contrast, ZF1 with its large hydrophobic plateau is able to destabilize the double-stranded regions adjacent to the guanines bound by ZF2. The linker residues and the internal dynamics of NC regulate therefore the different functions of the two zinc fingers that are required for an optimal chaperone activity.
Conflict of interest statement
Figures
Similar articles
-
The N-terminal zinc finger and flanking basic domains represent the minimal region of the human immunodeficiency virus type-1 nucleocapsid protein for targeting chaperone function.Biochemistry. 2013 Nov 19;52(46):8226-36. doi: 10.1021/bi401250a. Epub 2013 Nov 6. Biochemistry. 2013. PMID: 24144434 Free PMC article.
-
Subtle alterations of the native zinc finger structures have dramatic effects on the nucleic acid chaperone activity of human immunodeficiency virus type 1 nucleocapsid protein.J Virol. 2002 May;76(9):4370-8. doi: 10.1128/jvi.76.9.4370-4378.2002. J Virol. 2002. PMID: 11932404 Free PMC article.
-
A single zinc finger optimizes the DNA interactions of the nucleocapsid protein of the yeast retrotransposon Ty3.Nucleic Acids Res. 2012 Jan;40(2):751-60. doi: 10.1093/nar/gkr726. Epub 2011 Sep 14. Nucleic Acids Res. 2012. PMID: 21917850 Free PMC article.
-
Flexible nature and specific functions of the HIV-1 nucleocapsid protein.J Mol Biol. 2011 Jul 22;410(4):565-81. doi: 10.1016/j.jmb.2011.03.037. J Mol Biol. 2011. PMID: 21762801 Review.
-
Properties and functions of the nucleocapsid protein in virus assembly.RNA Biol. 2010 Nov-Dec;7(6):744-53. doi: 10.4161/rna.7.6.14065. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21157181 Free PMC article. Review.
Cited by
-
Metal cofactor modulated folding and target recognition of HIV-1 NCp7.PLoS One. 2018 May 1;13(5):e0196662. doi: 10.1371/journal.pone.0196662. eCollection 2018. PLoS One. 2018. PMID: 29715277 Free PMC article.
-
RNA Binding Suppresses Tsg101 Recognition of Ub-Modified Gag and Facilitates Recruitment to the Plasma Membrane.Viruses. 2020 Apr 15;12(4):447. doi: 10.3390/v12040447. Viruses. 2020. PMID: 32326417 Free PMC article.
-
Overview of the Nucleic-Acid Binding Properties of the HIV-1 Nucleocapsid Protein in Its Different Maturation States.Viruses. 2020 Sep 29;12(10):1109. doi: 10.3390/v12101109. Viruses. 2020. PMID: 33003650 Free PMC article. Review.
-
HIV-1 nucleocapsid and ESCRT-component Tsg101 interplay prevents HIV from turning into a DNA-containing virus.Nucleic Acids Res. 2015 Jan;43(1):336-47. doi: 10.1093/nar/gku1232. Epub 2014 Dec 8. Nucleic Acids Res. 2015. PMID: 25488808 Free PMC article.
-
Modulation of the HIV nucleocapsid dynamics finely tunes its RNA-binding properties during virion genesis.Nucleic Acids Res. 2018 Oct 12;46(18):9699-9710. doi: 10.1093/nar/gky612. Nucleic Acids Res. 2018. PMID: 29986076 Free PMC article.
References
-
- Levin JG, Guo J, Rouzina I, Musier-Forsyth K (2005) Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol 80: 217–286. - PubMed
-
- Darlix JL, Garrido JL, Morellet N, Mély Y, de Rocquigny H (2007) Properties, functions, and drug targeting of the multifunctional nucleocapsid protein of the human immunodeficiency virus. Adv Pharmacol 55: 299–346. - PubMed
-
- Darlix JL, Godet J, Ivanyi-Nagy R, Fossé P, Mauffret O, et al. (2011) Flexible Nature and Specific Functions of the HIV-1 Nucleocapsid Protein. J Mol Biol 410: 565–581. - PubMed
-
- Musier-Forsyth K (2010) RNA remodeling by chaperones and helicases. RNA Biol 7: 632–633. - PubMed
-
- Cristofari G, Darlix JL (2002) The ubiquitous nature of RNA chaperone proteins. Prog Nucleic Acid Res Mol Biol 72: 223–268. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

