Amoebozoa possess lineage-specific globin gene repertoires gained by individual horizontal gene transfers
- PMID: 25013378
- PMCID: PMC4081604
- DOI: 10.7150/ijbs.8327
Amoebozoa possess lineage-specific globin gene repertoires gained by individual horizontal gene transfers
Abstract
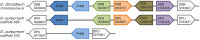
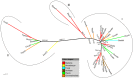
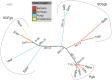
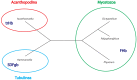
The Amoebozoa represent a clade of unicellular amoeboid organisms that display a wide variety of lifestyles, including free-living and parasitic species. For example, the social amoeba Dictyostelium discoideum has the ability to aggregate into a multicellular fruiting body upon starvation, while the pathogenic amoeba Entamoeba histolytica is a parasite of humans. Globins are small heme proteins that are present in almost all extant organisms. Although several genomes of amoebozoan species have been sequenced, little is known about the phyletic distribution of globin genes within this phylum. Only two flavohemoglobins (FHbs) of D. discoideum have been reported and characterized previously while the genomes of Entamoeba species are apparently devoid of globin genes. We investigated eleven amoebozoan species for the presence of globin genes by genomic and phylogenetic in silico analyses. Additional FHb genes were identified in the genomes of four social amoebas and the true slime mold Physarum polycephalum. Moreover, a single-domain globin (SDFgb) of Hartmannella vermiformis, as well as two truncated hemoglobins (trHbs) of Acanthamoeba castellanii were identified. Phylogenetic evidence suggests that these globin genes were independently acquired via horizontal gene transfer from some ancestral bacteria. Furthermore, the phylogenetic tree of amoebozoan FHbs indicates that they do not share a common ancestry and that a transfer of FHbs from bacteria to amoeba occurred multiple times.
Keywords: Amoebozoa; globin genes.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



Similar articles
-
Sampling gene diversity across the supergroup Amoebozoa: large EST data sets from Acanthamoeba castellanii, Hartmannella vermiformis, Physarum polycephalum, Hyperamoeba dachnaya and Hyperamoeba sp.Protist. 2008 Apr;159(2):269-81. doi: 10.1016/j.protis.2007.12.001. Epub 2008 Feb 13. Protist. 2008. PMID: 18276190
-
Abundant 5S rRNA-like transcripts encoded by the mitochondrial genome in amoebozoa.Eukaryot Cell. 2010 May;9(5):762-73. doi: 10.1128/EC.00013-10. Epub 2010 Mar 19. Eukaryot Cell. 2010. PMID: 20304999 Free PMC article.
-
Uncovering Cryptic Diversity in Two Amoebozoan Species Using Complete Mitochondrial Genome Sequences.J Eukaryot Microbiol. 2016 Jan-Feb;63(1):112-22. doi: 10.1111/jeu.12253. Epub 2015 Aug 28. J Eukaryot Microbiol. 2016. PMID: 26211788
-
Comparative genomics in the Amoebozoa clade.Biol Rev Camb Philos Soc. 2013 Feb;88(1):215-25. doi: 10.1111/j.1469-185X.2012.00248.x. Epub 2012 Nov 8. Biol Rev Camb Philos Soc. 2013. PMID: 23134060 Review.
-
Comparative genomics of Dictyostelium discoideum and Entamoeba histolytica.Curr Opin Microbiol. 2005 Oct;8(5):606-11. doi: 10.1016/j.mib.2005.08.009. Curr Opin Microbiol. 2005. PMID: 16125444 Review.
Cited by
-
The aerotaxis of Dictyostelium discoideum is independent of mitochondria, nitric oxide and oxidative stress.Front Cell Dev Biol. 2023 Jun 15;11:1134011. doi: 10.3389/fcell.2023.1134011. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37397260 Free PMC article.
-
CCA-Addition Gone Wild: Unusual Occurrence and Phylogeny of Four Different tRNA Nucleotidyltransferases in Acanthamoeba castellanii.Mol Biol Evol. 2021 Mar 9;38(3):1006-1017. doi: 10.1093/molbev/msaa270. Mol Biol Evol. 2021. PMID: 33095240 Free PMC article.
-
Evolutionary analysis of globin domains from kinetoplastids.Arch Microbiol. 2022 Jul 16;204(8):493. doi: 10.1007/s00203-022-03107-1. Arch Microbiol. 2022. PMID: 35841431
References
-
- Vinogradov S, Hoogewijs D, Vanfleteren J, Dewilde S, Moens L, Hankeln T. Evolution of the globin superfamily and its function. In Homoglobin: Recent Developments and Topics. Edited by Nagai M. Kerala, IND: Research Signpost; 2011. pp. 231–254.
-
- Vinogradov SN, Moens L. Diversity of globin function: enzymatic, transport, storage, and sensing. The Journal of biological chemistry. 2008;283:8773–8777. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

