An inside job: how endosomal Na(+)/H(+) exchangers link to autism and neurological disease
- PMID: 25002837
- PMCID: PMC4066934
- DOI: 10.3389/fncel.2014.00172
An inside job: how endosomal Na(+)/H(+) exchangers link to autism and neurological disease
Abstract
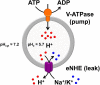
Autism imposes a major impediment to childhood development and a huge emotional and financial burden on society. In recent years, there has been rapidly accumulating genetic evidence that links the eNHE, a subset of Na(+)/H(+) exchangers that localize to intracellular vesicles, to a variety of neurological conditions including autism, attention deficit hyperactivity disorder (ADHD), intellectual disability, and epilepsy. By providing a leak pathway for protons pumped by the V-ATPase, eNHE determine luminal pH and regulate cation (Na(+), K(+)) content in early and recycling endosomal compartments. Loss-of-function mutations in eNHE cause hyperacidification of endosomal lumen, as a result of imbalance in pump and leak pathways. Two isoforms, NHE6 and NHE9 are highly expressed in brain, including hippocampus and cortex. Here, we summarize evidence for the importance of luminal cation content and pH on processing, delivery and fate of cargo. Drawing upon insights from model organisms and mammalian cells we show how eNHE affect surface expression and function of membrane receptors and neurotransmitter transporters. These studies lead to cellular models of eNHE activity in pre- and post-synaptic neurons and astrocytes, where they could impact synapse development and plasticity. The study of eNHE has provided new insight on the mechanism of autism and other debilitating neurological disorders and opened up new possibilities for therapeutic intervention.
Keywords: ADHD; Christianson syndrome; SLC9A6; SLC9A9; autism; endosomes; sodium proton exchanger; trafficking.
Figures

Similar articles
-
A potential gain-of-function variant of SLC9A6 leads to endosomal alkalinization and neuronal atrophy associated with Christianson Syndrome.Neurobiol Dis. 2019 Jan;121:187-204. doi: 10.1016/j.nbd.2018.10.002. Epub 2018 Oct 5. Neurobiol Dis. 2019. PMID: 30296617
-
Loss of Christianson Syndrome Na+/H+ Exchanger 6 (NHE6) Causes Abnormal Endosome Maturation and Trafficking Underlying Lysosome Dysfunction in Neurons.J Neurosci. 2021 Nov 3;41(44):9235-9256. doi: 10.1523/JNEUROSCI.1244-20.2021. Epub 2021 Sep 15. J Neurosci. 2021. PMID: 34526390 Free PMC article.
-
Assorted dysfunctions of endosomal alkali cation/proton exchanger SLC9A6 variants linked to Christianson syndrome.J Biol Chem. 2020 May 15;295(20):7075-7095. doi: 10.1074/jbc.RA120.012614. Epub 2020 Apr 10. J Biol Chem. 2020. PMID: 32277048 Free PMC article.
-
Roles of Endomembrane Alkali Cation/Proton Exchangers in Synaptic Function and Neurodevelopmental Disorders.Front Physiol. 2022 Apr 25;13:892196. doi: 10.3389/fphys.2022.892196. eCollection 2022. Front Physiol. 2022. PMID: 35547574 Free PMC article. Review.
-
Emerging links between endosomal pH and cancer.Cancer Metastasis Rev. 2020 Jun;39(2):519-534. doi: 10.1007/s10555-020-09870-1. Cancer Metastasis Rev. 2020. PMID: 32253638 Free PMC article. Review.
Cited by
-
Exonic deletion of SLC9A9 in autism with epilepsy.Neurol Genet. 2016 Feb 25;2(2):e62. doi: 10.1212/NXG.0000000000000062. eCollection 2016 Apr. Neurol Genet. 2016. PMID: 27123481 Free PMC article.
-
Loss of endosomal exchanger NHE6 leads to pathological changes in tau in human neurons.Stem Cell Reports. 2022 Sep 13;17(9):2111-2126. doi: 10.1016/j.stemcr.2022.08.001. Epub 2022 Sep 1. Stem Cell Reports. 2022. PMID: 36055242 Free PMC article.
-
Pathophysiology of hepatic Na+/H+ exchange (Review).Exp Ther Med. 2020 Aug;20(2):1220-1229. doi: 10.3892/etm.2020.8888. Epub 2020 Jun 12. Exp Ther Med. 2020. PMID: 32742358 Free PMC article. Review.
-
Neuron membrane trafficking and protein kinases involved in autism and ADHD.Int J Mol Sci. 2015 Jan 30;16(2):3095-115. doi: 10.3390/ijms16023095. Int J Mol Sci. 2015. PMID: 25647412 Free PMC article. Review.
-
Phenotypic and genetic spectrum of ATP6V1A encephalopathy: a disorder of lysosomal homeostasis.Brain. 2022 Aug 27;145(8):2687-2703. doi: 10.1093/brain/awac145. Brain. 2022. PMID: 35675510 Free PMC article.
References
-
- Amr M., Raddad D., El-Mehesh F., Bakr A., Sallam K., Amin T. (2012). Comorbid psychiatric disorders in Arab children with autism spectrum disorders. Res. Autism Spectr. Disord. 6, 240–248 10.1016/j.rasd.2011.05.005 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

