Accessory proteins of SARS-CoV and other coronaviruses
- PMID: 24995382
- PMCID: PMC7113789
- DOI: 10.1016/j.antiviral.2014.06.013
Accessory proteins of SARS-CoV and other coronaviruses
Abstract
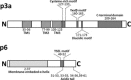
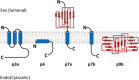
The huge RNA genome of SARS coronavirus comprises a number of open reading frames that code for a total of eight accessory proteins. Although none of these are essential for virus replication, some appear to have a role in virus pathogenesis. Notably, some SARS-CoV accessory proteins have been shown to modulate the interferon signaling pathways and the production of pro-inflammatory cytokines. The structural information on these proteins is also limited, with only two (p7a and p9b) having their structures determined by X-ray crystallography. This review makes an attempt to summarize the published knowledge on SARS-CoV accessory proteins, with an emphasis on their involvement in virus-host interaction. The accessory proteins of other coronaviruses are also briefly discussed. This paper forms part of a series of invited articles in Antiviral Research on "From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses" (see Introduction by Hilgenfeld and Peiris (2013)).
Keywords: Accessory proteins; Other coronaviruses; SARS-CoV; Structure and function.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures



Similar articles
-
Characterization of accessory genes in coronavirus genomes.Virol J. 2020 Aug 27;17(1):131. doi: 10.1186/s12985-020-01402-1. Virol J. 2020. PMID: 32854725 Free PMC article.
-
The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis.Viruses. 2012 Nov 7;4(11):2902-23. doi: 10.3390/v4112902. Viruses. 2012. PMID: 23202509 Free PMC article. Review.
-
The N-Terminal Region of Middle East Respiratory Syndrome Coronavirus Accessory Protein 8b Is Essential for Enhanced Virulence of an Attenuated Murine Coronavirus.J Virol. 2022 Feb 9;96(3):e0184221. doi: 10.1128/JVI.01842-21. Epub 2021 Nov 24. J Virol. 2022. PMID: 34817197 Free PMC article.
-
Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage.J Mol Biol. 2003 Aug 29;331(5):991-1004. doi: 10.1016/s0022-2836(03)00865-9. J Mol Biol. 2003. PMID: 12927536 Free PMC article.
-
The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds.Antiviral Res. 2015 Mar;115:21-38. doi: 10.1016/j.antiviral.2014.12.015. Epub 2014 Dec 29. Antiviral Res. 2015. PMID: 25554382 Free PMC article. Review.
Cited by
-
SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5-MAVS, TLR3-TRIF, and cGAS-STING signaling pathways.J Med Virol. 2021 Sep;93(9):5376-5389. doi: 10.1002/jmv.27050. Epub 2021 May 9. J Med Virol. 2021. PMID: 33913550 Free PMC article.
-
Epidemiology of coronaviruses, genetics, vaccines, and scenario of current pandemic of coronavirus diseases 2019 (COVID-19): a fuzzy set approach.Hum Vaccin Immunother. 2021 May 4;17(5):1296-1303. doi: 10.1080/21645515.2020.1798697. Epub 2021 Mar 15. Hum Vaccin Immunother. 2021. PMID: 33720797 Free PMC article. Review.
-
SARS-CoV-2 gene content and COVID-19 mutation impact by comparing 44 Sarbecovirus genomes.Nat Commun. 2021 May 11;12(1):2642. doi: 10.1038/s41467-021-22905-7. Nat Commun. 2021. PMID: 33976134 Free PMC article.
-
Middle East Respiratory Coronavirus Accessory Protein 4a Inhibits PKR-Mediated Antiviral Stress Responses.PLoS Pathog. 2016 Oct 26;12(10):e1005982. doi: 10.1371/journal.ppat.1005982. eCollection 2016 Oct. PLoS Pathog. 2016. PMID: 27783669 Free PMC article.
-
Potential Anti-Coronavirus Agents and the Pharmacologic Mechanisms.Drug Des Devel Ther. 2021 Mar 17;15:1213-1223. doi: 10.2147/DDDT.S293216. eCollection 2021. Drug Des Devel Ther. 2021. PMID: 33762818 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

