The dissolution of double Holliday junctions
- PMID: 24984776
- PMCID: PMC4067992
- DOI: 10.1101/cshperspect.a016477
The dissolution of double Holliday junctions
Abstract
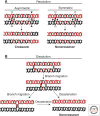
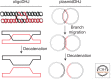
Double Holliday junctions (dHJS) are important intermediates of homologous recombination. The separate junctions can each be cleaved by DNA structure-selective endonucleases known as Holliday junction resolvases. Alternatively, double Holliday junctions can be processed by a reaction known as "double Holliday junction dissolution." This reaction requires the cooperative action of a so-called "dissolvasome" comprising a Holliday junction branch migration enzyme (Sgs1/BLM RecQ helicase) and a type IA topoisomerase (Top3/TopoIIIα) in complex with its OB (oligonucleotide/oligosaccharide binding) fold containing accessory factor (Rmi1). This review details our current knowledge of the dissolution process and the players involved in catalyzing this mechanistically complex means of completing homologous recombination reactions.
Copyright © 2014 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures
Similar articles
-
Dissolution of double Holliday junctions by the concerted action of BLM and topoisomerase IIIalpha.Methods Mol Biol. 2009;582:91-102. doi: 10.1007/978-1-60761-340-4_8. Methods Mol Biol. 2009. PMID: 19763944
-
BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates.Proc Natl Acad Sci U S A. 2006 Mar 14;103(11):4068-73. doi: 10.1073/pnas.0508295103. Epub 2006 Mar 6. Proc Natl Acad Sci U S A. 2006. PMID: 16537486 Free PMC article.
-
Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3.Nat Struct Mol Biol. 2010 Nov;17(11):1377-82. doi: 10.1038/nsmb.1919. Epub 2010 Oct 10. Nat Struct Mol Biol. 2010. PMID: 20935631 Free PMC article.
-
Resolution of Recombination Intermediates: Mechanisms and Regulation.Cold Spring Harb Symp Quant Biol. 2015;80:103-9. doi: 10.1101/sqb.2015.80.027649. Epub 2015 Sep 14. Cold Spring Harb Symp Quant Biol. 2015. PMID: 26370409 Review.
-
A Structural Guide to the Bloom Syndrome Complex.Structure. 2021 Feb 4;29(2):99-113. doi: 10.1016/j.str.2020.11.020. Epub 2020 Dec 22. Structure. 2021. PMID: 33357470 Review.
Cited by
-
Helicase Mechanisms During Homologous Recombination in Saccharomyces cerevisiae.Annu Rev Biophys. 2019 May 6;48:255-273. doi: 10.1146/annurev-biophys-052118-115418. Epub 2019 Mar 11. Annu Rev Biophys. 2019. PMID: 30857400 Free PMC article. Review.
-
Age-Dependent Alterations in Meiotic Recombination Cause Chromosome Segregation Errors in Spermatocytes.Cell. 2017 Oct 19;171(3):601-614.e13. doi: 10.1016/j.cell.2017.08.042. Epub 2017 Sep 21. Cell. 2017. PMID: 28942922 Free PMC article.
-
Structure and Function of a Novel ATPase that Interacts with Holliday Junction Resolvase Hjc and Promotes Branch Migration.J Mol Biol. 2017 Apr 7;429(7):1009-1029. doi: 10.1016/j.jmb.2017.02.016. Epub 2017 Feb 24. J Mol Biol. 2017. PMID: 28238763 Free PMC article.
-
ATRX and RECQ5 define distinct homologous recombination subpathways.Proc Natl Acad Sci U S A. 2021 Jan 19;118(3):e2010370118. doi: 10.1073/pnas.2010370118. Proc Natl Acad Sci U S A. 2021. PMID: 33431668 Free PMC article.
-
The Bloom syndrome complex senses RPA-coated single-stranded DNA to restart stalled replication forks.Nat Commun. 2021 Jan 26;12(1):585. doi: 10.1038/s41467-020-20818-5. Nat Commun. 2021. PMID: 33500419 Free PMC article.
References
-
- Bachrati CZ, Hickson ID 2009. Dissolution of double Holliday junctions by the concerted action of BLM and topoisomerase IIIα. Methods Mol Biol 582: 91–102 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
