Insulator function and topological domain border strength scale with architectural protein occupancy
- PMID: 24981874
- PMCID: PMC4226948
- DOI: 10.1186/gb-2014-15-5-r82
Insulator function and topological domain border strength scale with architectural protein occupancy
Abstract
Background: Chromosome conformation capture studies suggest that eukaryotic genomes are organized into structures called topologically associating domains. The borders of these domains are highly enriched for architectural proteins with characterized roles in insulator function. However, a majority of architectural protein binding sites localize within topological domains, suggesting sites associated with domain borders represent a functionally different subclass of these regulatory elements. How topologically associating domains are established and what differentiates border-associated from non-border architectural protein binding sites remain unanswered questions.
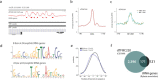
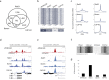
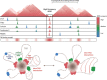
Results: By mapping the genome-wide target sites for several Drosophila architectural proteins, including previously uncharacterized profiles for TFIIIC and SMC-containing condensin complexes, we uncover an extensive pattern of colocalization in which architectural proteins establish dense clusters at the borders of topological domains. Reporter-based enhancer-blocking insulator activity as well as endogenous domain border strength scale with the occupancy level of architectural protein binding sites, suggesting co-binding by architectural proteins underlies the functional potential of these loci. Analyses in mouse and human stem cells suggest that clustering of architectural proteins is a general feature of genome organization, and conserved architectural protein binding sites may underlie the tissue-invariant nature of topologically associating domains observed in mammals.
Conclusions: We identify a spectrum of architectural protein occupancy that scales with the topological structure of chromosomes and the regulatory potential of these elements. Whereas high occupancy architectural protein binding sites associate with robust partitioning of topologically associating domains and robust insulator function, low occupancy sites appear reserved for gene-specific regulation within topological domains.
Figures



Comment in
-
Binding the boundaries of chromatin domains.Genome Biol. 2014 Jun 30;15(6):121. doi: 10.1186/gb4183. Genome Biol. 2014. PMID: 25000862 Free PMC article.
Similar articles
-
Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders.Genome Biol. 2016 Aug 31;17(1):182. doi: 10.1186/s13059-016-1043-8. Genome Biol. 2016. PMID: 27582050 Free PMC article.
-
Insulators and domains of gene expression.Curr Opin Genet Dev. 2016 Apr;37:17-26. doi: 10.1016/j.gde.2015.11.009. Epub 2016 Jan 20. Curr Opin Genet Dev. 2016. PMID: 26802288 Review.
-
Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains.Genome Res. 2012 Nov;22(11):2176-87. doi: 10.1101/gr.136788.111. Epub 2012 Jun 21. Genome Res. 2012. PMID: 22722341 Free PMC article.
-
Sub-kb Hi-C in D. melanogaster reveals conserved characteristics of TADs between insect and mammalian cells.Nat Commun. 2018 Jan 15;9(1):188. doi: 10.1038/s41467-017-02526-9. Nat Commun. 2018. PMID: 29335463 Free PMC article.
-
Chromatin Architecture in the Fly: Living without CTCF/Cohesin Loop Extrusion?: Alternating Chromatin States Provide a Basis for Domain Architecture in Drosophila.Bioessays. 2019 Sep;41(9):e1900048. doi: 10.1002/bies.201900048. Epub 2019 Jul 1. Bioessays. 2019. PMID: 31264253 Review.
Cited by
-
ZSCAN5B and primate-specific paralogs bind RNA polymerase III genes and extra-TFIIIC (ETC) sites to modulate mitotic progression.Oncotarget. 2016 Nov 8;7(45):72571-72592. doi: 10.18632/oncotarget.12508. Oncotarget. 2016. PMID: 27732952 Free PMC article.
-
Characterization of Tau95 led to the identification of a four-subunit TFIIIC complex in trypanosomatid parasites.Appl Microbiol Biotechnol. 2024 Dec;108(1):109. doi: 10.1007/s00253-023-12903-8. Epub 2024 Jan 10. Appl Microbiol Biotechnol. 2024. PMID: 38204130 Free PMC article.
-
Balancing up and downregulation of the C. elegans X chromosomes.Curr Opin Genet Dev. 2015 Apr;31:50-6. doi: 10.1016/j.gde.2015.04.001. Epub 2015 May 16. Curr Opin Genet Dev. 2015. PMID: 25966908 Free PMC article. Review.
-
Phosphorylated histone variant γH2Av is associated with chromatin insulators in Drosophila.PLoS Genet. 2022 Oct 5;18(10):e1010396. doi: 10.1371/journal.pgen.1010396. eCollection 2022 Oct. PLoS Genet. 2022. PMID: 36197938 Free PMC article.
-
Taking cohesin and condensin in context.PLoS Genet. 2018 Jan 25;14(1):e1007118. doi: 10.1371/journal.pgen.1007118. eCollection 2018 Jan. PLoS Genet. 2018. PMID: 29370184 Free PMC article. Review.
References
-
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. - PubMed
-
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, Bland MJ, Wagstaff W, Dalton S, McDevitt TC, Sen R, Dekker J, Taylor J, Corces VG. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. - PMC - PubMed
-
- Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, Ing-Simmons E, Lenhard B, Giorgetti L, Heard E, Fisher AG, Flicek P, Dekker J, Merkenschlager M. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013;23:2066–2077. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

