Mechanisms underlying mutational signatures in human cancers
- PMID: 24981601
- PMCID: PMC6044419
- DOI: 10.1038/nrg3729
Mechanisms underlying mutational signatures in human cancers
Abstract
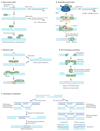
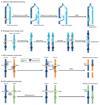
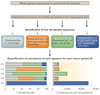
The collective somatic mutations observed in a cancer are the outcome of multiple mutagenic processes that have been operative over the lifetime of a patient. Each process leaves a characteristic imprint--a mutational signature--on the cancer genome, which is defined by the type of DNA damage and DNA repair processes that result in base substitutions, insertions and deletions or structural variations. With the advent of whole-genome sequencing, researchers are identifying an increasing array of these signatures. Mutational signatures can be used as a physiological readout of the biological history of a cancer and also have potential use for discerning ongoing mutational processes from historical ones, thus possibly revealing new targets for anticancer therapies.
Conflict of interest statement
The authors declare no competing interests.
Figures




Comment in
-
Cancer genomics: A catalogue of somatic mutations.Nat Rev Genet. 2016 Jul;17(7):378. doi: 10.1038/nrg.2016.65. Epub 2016 May 9. Nat Rev Genet. 2016. PMID: 27156977 No abstract available.
Similar articles
-
Mutational spectra and mutational signatures: Insights into cancer aetiology and mechanisms of DNA damage and repair.DNA Repair (Amst). 2018 Nov;71:6-11. doi: 10.1016/j.dnarep.2018.08.003. Epub 2018 Aug 24. DNA Repair (Amst). 2018. PMID: 30236628 Free PMC article. Review.
-
MutationalPatterns: the one stop shop for the analysis of mutational processes.BMC Genomics. 2022 Feb 15;23(1):134. doi: 10.1186/s12864-022-08357-3. BMC Genomics. 2022. PMID: 35168570 Free PMC article.
-
Deciphering signatures of mutational processes operative in human cancer.Cell Rep. 2013 Jan 31;3(1):246-59. doi: 10.1016/j.celrep.2012.12.008. Epub 2013 Jan 10. Cell Rep. 2013. PMID: 23318258 Free PMC article.
-
Computational approaches for discovery of mutational signatures in cancer.Brief Bioinform. 2019 Jan 18;20(1):77-88. doi: 10.1093/bib/bbx082. Brief Bioinform. 2019. PMID: 28968631 Free PMC article. Review.
-
The repertoire of mutational signatures in human cancer.Nature. 2020 Feb;578(7793):94-101. doi: 10.1038/s41586-020-1943-3. Epub 2020 Feb 5. Nature. 2020. PMID: 32025018 Free PMC article.
Cited by
-
Mutation signatures implicate aristolochic acid in bladder cancer development.Genome Med. 2015 Apr 28;7(1):38. doi: 10.1186/s13073-015-0161-3. eCollection 2015. Genome Med. 2015. PMID: 26015808 Free PMC article.
-
Mutation Processes in 293-Based Clones Overexpressing the DNA Cytosine Deaminase APOBEC3B.PLoS One. 2016 May 10;11(5):e0155391. doi: 10.1371/journal.pone.0155391. eCollection 2016. PLoS One. 2016. PMID: 27163364 Free PMC article.
-
Base changes in tumour DNA have the power to reveal the causes and evolution of cancer.Oncogene. 2017 Jan 12;36(2):158-167. doi: 10.1038/onc.2016.192. Epub 2016 Jun 6. Oncogene. 2017. PMID: 27270430 Free PMC article. Review.
-
Genomic Characteristics and the Potential Clinical Implications in Oligometastatic Non-Small Cell Lung Cancer.Cancer Res Treat. 2023 Jul;55(3):814-831. doi: 10.4143/crt.2022.1315. Epub 2023 Jan 12. Cancer Res Treat. 2023. PMID: 36634615 Free PMC article.
-
Multiple interaction nodes define the postreplication repair response to UV-induced DNA damage that is defective in melanomas and correlated with UV signature mutation load.Mol Oncol. 2020 Jan;14(1):22-41. doi: 10.1002/1878-0261.12601. Epub 2019 Dec 19. Mol Oncol. 2020. PMID: 31733171 Free PMC article.
References
-
- Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. [This study shows the prevalence of somatic mutations in human cancer genomes, which indicates that most of the mutations do not drive oncogenesis. Nevertheless, it provides evidence for driver mutations that are actively involved in tumour development.] - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

