Population distribution analyses reveal a hierarchy of molecular players underlying parallel endocytic pathways
- PMID: 24971745
- PMCID: PMC4074053
- DOI: 10.1371/journal.pone.0100554
Population distribution analyses reveal a hierarchy of molecular players underlying parallel endocytic pathways
Erratum in
-
Correction: Population Distribution Analyses Reveal a Hierarchy of Molecular Players Underlying Parallel Endocytic Pathways.PLoS One. 2018 Sep 21;13(9):e0204770. doi: 10.1371/journal.pone.0204770. eCollection 2018. PLoS One. 2018. PMID: 30240414 Free PMC article.
Abstract
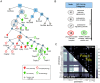
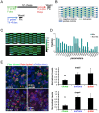
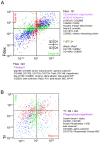
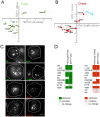
Single-cell-resolved measurements reveal heterogeneous distributions of clathrin-dependent (CD) and -independent (CLIC/GEEC: CG) endocytic activity in Drosophila cell populations. dsRNA-mediated knockdown of core versus peripheral endocytic machinery induces strong changes in the mean, or subtle changes in the shapes of these distributions, respectively. By quantifying these subtle shape changes for 27 single-cell features which report on endocytic activity and cell morphology, we organize 1072 Drosophila genes into a tree-like hierarchy. We find that tree nodes contain gene sets enriched in functional classes and protein complexes, providing a portrait of core and peripheral control of CD and CG endocytosis. For 470 genes we obtain additional features from separate assays and classify them into early- or late-acting genes of the endocytic pathways. Detailed analyses of specific genes at intermediate levels of the tree suggest that Vacuolar ATPase and lysosomal genes involved in vacuolar biogenesis play an evolutionarily conserved role in CG endocytosis.
Conflict of interest statement
Figures


Similar articles
-
Endocytic crosstalk: cavins, caveolins, and caveolae regulate clathrin-independent endocytosis.PLoS Biol. 2014 Apr 8;12(4):e1001832. doi: 10.1371/journal.pbio.1001832. eCollection 2014 Apr. PLoS Biol. 2014. PMID: 24714042 Free PMC article.
-
Shibire mutations reveal distinct dynamin-independent and -dependent endocytic pathways in primary cultures of Drosophila hemocytes.J Cell Sci. 2003 Aug 15;116(Pt 16):3373-86. doi: 10.1242/jcs.00637. J Cell Sci. 2003. PMID: 12857788
-
The DISABLED protein functions in CLATHRIN-mediated synaptic vesicle endocytosis and exoendocytic coupling at the active zone.Proc Natl Acad Sci U S A. 2011 Jun 21;108(25):E222-9. doi: 10.1073/pnas.1102231108. Epub 2011 May 23. Proc Natl Acad Sci U S A. 2011. PMID: 21606364 Free PMC article.
-
Molecular mechanisms of clathrin-independent endocytosis.J Cell Sci. 2009 Jun 1;122(Pt 11):1713-21. doi: 10.1242/jcs.033951. J Cell Sci. 2009. PMID: 19461071 Free PMC article. Review.
-
Mechanisms of endocytosis.Annu Rev Biochem. 2009;78:857-902. doi: 10.1146/annurev.biochem.78.081307.110540. Annu Rev Biochem. 2009. PMID: 19317650 Review.
Cited by
-
Recent advances in clathrin-independent endocytosis.F1000Res. 2019 Jan 31;8:F1000 Faculty Rev-138. doi: 10.12688/f1000research.16549.1. eCollection 2019. F1000Res. 2019. PMID: 30774931 Free PMC article. Review.
-
Mechanochemical feedback control of dynamin independent endocytosis modulates membrane tension in adherent cells.Nat Commun. 2018 Oct 11;9(1):4217. doi: 10.1038/s41467-018-06738-5. Nat Commun. 2018. PMID: 30310066 Free PMC article.
-
Spoiled for Choice: Diverse Endocytic Pathways Function at the Cell Surface.Annu Rev Cell Dev Biol. 2019 Oct 6;35:55-84. doi: 10.1146/annurev-cellbio-100617-062710. Epub 2019 Jul 5. Annu Rev Cell Dev Biol. 2019. PMID: 31283376 Free PMC article. Review.
-
Strategies to target SARS-CoV-2 entry and infection using dual mechanisms of inhibition by acidification inhibitors.PLoS Pathog. 2021 Jul 12;17(7):e1009706. doi: 10.1371/journal.ppat.1009706. eCollection 2021 Jul. PLoS Pathog. 2021. PMID: 34252168 Free PMC article.
-
SUMOylation of EHD3 Modulates Tubulation of the Endocytic Recycling Compartment.PLoS One. 2015 Jul 30;10(7):e0134053. doi: 10.1371/journal.pone.0134053. eCollection 2015. PLoS One. 2015. PMID: 26226295 Free PMC article.
References
-
- Mayor S, Pagano RE (2007) Pathways of clathrin-independent endocytosis. Nat Rev Mol Cell Biol 8: 603–612. - PubMed
-
- Doherty GJ, McMahon HT (2009) Mechanisms of endocytosis. Annual review of biochemistry 78: 857–902. - PubMed
-
- Howes MT, Mayor S, Parton RG (2010) Molecules, mechanisms, and cellular roles of clathrin-independent endocytosis. Current opinion in cell biology 22: 519–527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

