Growth factor and Th2 cytokine signaling pathways converge at STAT6 to promote arginase expression in progressive experimental visceral leishmaniasis
- PMID: 24967908
- PMCID: PMC4072777
- DOI: 10.1371/journal.ppat.1004165
Growth factor and Th2 cytokine signaling pathways converge at STAT6 to promote arginase expression in progressive experimental visceral leishmaniasis
Abstract
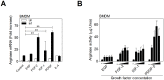
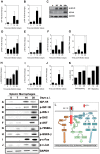
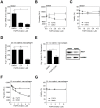
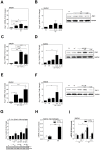
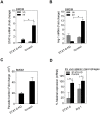
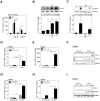
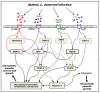
Host arginase 1 (arg1) expression is a significant contributor to the pathogenesis of progressive visceral leishmaniasis (VL), a neglected tropical disease caused by the intracellular protozoan Leishmania donovani. Previously we found that parasite-induced arg1 expression in macrophages was dependent on STAT6 activation. Arg1 expression was amplified by, but did not require, IL-4, and required de novo synthesis of unknown protein(s). To further explore the mechanisms involved in arg1 regulation in VL, we screened a panel of kinase inhibitors and found that inhibitors of growth factor signaling reduced arg1 expression in splenic macrophages from hamsters with VL. Analysis of growth factors and their signaling pathways revealed that the Fibroblast Growth Factor Receptor 1 (FGFR-1) and Insulin-like Growth Factor 1 Receptor (IGF-1R) and a number of downstream signaling proteins were activated in splenic macrophages isolated from hamsters infected with L. donovani. Recombinant FGF-2 and IGF-1 increased the expression of arg1 in L. donovani infected hamster macrophages, and this induction was augmented by IL-4. Inhibition of FGFR-1 and IGF-1R decreased arg1 expression and restricted L. donovani replication in both in vitro and ex vivo models of infection. Inhibition of the downstream signaling molecules JAK and AKT also reduced the expression of arg1 in infected macrophages. STAT6 was activated in infected macrophages exposed to either FGF-2 or IGF-1, and STAT6 was critical to the FGFR-1- and IGF-1R-mediated expression of arg1. The converse was also true as inhibition of FGFR-1 and IGF-1R reduced the activation of STAT6 in infected macrophages. Collectively, these data indicate that the FGFR/IGF-1R and IL-4 signaling pathways converge at STAT6 to promote pathologic arg1 expression and intracellular parasite survival in VL. Targeted interruption of these pathological processes offers an approach to restrain this relentlessly progressive disease.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Splenic CD4+ T Cells in Progressive Visceral Leishmaniasis Show a Mixed Effector-Regulatory Phenotype and Impair Macrophage Effector Function through Inhibitory Receptor Expression.PLoS One. 2017 Jan 19;12(1):e0169496. doi: 10.1371/journal.pone.0169496. eCollection 2017. PLoS One. 2017. PMID: 28103263 Free PMC article.
-
Progressive visceral leishmaniasis is driven by dominant parasite-induced STAT6 activation and STAT6-dependent host arginase 1 expression.PLoS Pathog. 2012 Jan;8(1):e1002417. doi: 10.1371/journal.ppat.1002417. Epub 2012 Jan 19. PLoS Pathog. 2012. PMID: 22275864 Free PMC article.
-
Transcriptional Profiling in Experimental Visceral Leishmaniasis Reveals a Broad Splenic Inflammatory Environment that Conditions Macrophages toward a Disease-Promoting Phenotype.PLoS Pathog. 2017 Jan 31;13(1):e1006165. doi: 10.1371/journal.ppat.1006165. eCollection 2017 Jan. PLoS Pathog. 2017. PMID: 28141856 Free PMC article.
-
Neutrophils and Visceral Leishmaniasis: Impact on innate immune response and cross-talks with macrophages and dendritic cells.J Cell Physiol. 2021 Apr;236(4):2255-2267. doi: 10.1002/jcp.30029. Epub 2020 Dec 20. J Cell Physiol. 2021. PMID: 33345353 Review.
-
Pleiotropic Effect of Hormone Insulin-Like Growth Factor-I in Immune Response and Pathogenesis in Leishmaniases.J Immunol Res. 2021 May 4;2021:6614475. doi: 10.1155/2021/6614475. eCollection 2021. J Immunol Res. 2021. PMID: 34036108 Free PMC article. Review.
Cited by
-
Ghrelin regulating liver activity and its potential effects on liver fibrosis and Echinococcosis.Front Cell Infect Microbiol. 2024 Jan 8;13:1324134. doi: 10.3389/fcimb.2023.1324134. eCollection 2023. Front Cell Infect Microbiol. 2024. PMID: 38259969 Free PMC article. Review.
-
Splenic CD4+ T Cells in Progressive Visceral Leishmaniasis Show a Mixed Effector-Regulatory Phenotype and Impair Macrophage Effector Function through Inhibitory Receptor Expression.PLoS One. 2017 Jan 19;12(1):e0169496. doi: 10.1371/journal.pone.0169496. eCollection 2017. PLoS One. 2017. PMID: 28103263 Free PMC article.
-
Evaluation of In vitro and In vivo Protective Efficacy of Bauhinia variegata Against Leishmania donovani in Murine Model.Acta Parasitol. 2021 Sep;66(3):812-826. doi: 10.1007/s11686-020-00326-8. Epub 2021 Feb 2. Acta Parasitol. 2021. PMID: 33528770
-
In-situ proliferation contributes to the accumulation of myeloid cells in the spleen during progressive experimental visceral leishmaniasis.PLoS One. 2020 Nov 12;15(11):e0242337. doi: 10.1371/journal.pone.0242337. eCollection 2020. PLoS One. 2020. PMID: 33180876 Free PMC article.
-
Modulation of Macrophage Immunometabolism: A New Approach to Fight Infections.Front Immunol. 2022 Jan 26;13:780839. doi: 10.3389/fimmu.2022.780839. eCollection 2022. Front Immunol. 2022. PMID: 35154105 Free PMC article. Review.
References
-
- Green SJ, Nacy CA, Meltzer MS (1991) Cytokine-induced synthesis of nitrogen oxides in macrophages: a protective host response to Leishmania and other intracellular pathogens. J Leukoc Biol 50: 93–103. - PubMed
-
- Liew FY, Li Y, Moss D, Parkinson C, Rogers MV, et al. (1991) Resistance to Leishmania major infection correlates with the induction of nitric oxide synthase in murine macrophages. Eur J Immunol 21: 3009–3014. - PubMed
-
- Gordon S, Martinez FO (2010) Alternative activation of macrophages: mechanism and functions. Immunity 32: 593–604. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

