Tc1-like transposable elements in plant genomes
- PMID: 24926322
- PMCID: PMC4054914
- DOI: 10.1186/1759-8753-5-17
Tc1-like transposable elements in plant genomes
Abstract
Background: The Tc1/mariner superfamily of transposable elements (TEs) is widespread in animal genomes. Mariner-like elements, which bear a DDD triad catalytic motif, have been identified in a wide range of flowering plant species. However, as the founding member of the superfamily, Tc1-like elements that bear a DD34E triad catalytic motif are only known to unikonts (animals, fungi, and Entamoeba).
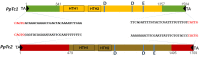
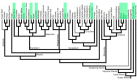
Results: Here we report the identification of Tc1-like elements (TLEs) in plant genomes. These elements bear the four terminal nucleotides and the characteristic DD34E triad motif of Tc1 element. The two TLE families (PpTc1, PpTc2) identified in the moss (Physcomitrella patens) genome contain highly similar copies. Multiple copies of PpTc1 are actively transcribed and the transcripts encode intact full length transposase coding sequences. TLEs are also found in angiosperm genome sequence databases of rice (Oryza sativa), dwarf birch (Betula nana), cabbage (Brassica rapa), hemp (Cannabis sativa), barley (Hordium valgare), lettuce (Lactuta sativa), poplar (Populus trichocarpa), pear (Pyrus x bretschneideri), and wheat (Triticum urartu).
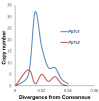
Conclusions: This study extends the occurrence of TLEs to the plant phylum. The elements in the moss genome have amplified recently and may still be capable of transposition. The TLEs are also present in angiosperm genomes, but apparently much less abundant than in moss.
Keywords: Evolution; Mariner-like elements; Moss; Plant genome; Tc1-like elements; Tc1-mariner-IS630 superfamily; Transposable elements; Transposition activity.
Figures




Similar articles
-
An Analysis of IS630/Tc1/mariner Transposons in the Genome of a Pacific Oyster, Crassostrea gigas.J Mol Evol. 2018 Oct;86(8):566-580. doi: 10.1007/s00239-018-9868-2. Epub 2018 Oct 3. J Mol Evol. 2018. PMID: 30283979
-
Gambol and Tc1 are two distinct families of DD34E transposons: analysis of the Anopheles gambiae genome expands the diversity of the IS630-Tc1-mariner superfamily.Insect Mol Biol. 2005 Oct;14(5):537-46. doi: 10.1111/j.1365-2583.2005.00584.x. Insect Mol Biol. 2005. PMID: 16164609
-
Phylogenetic analysis of the Tc1/mariner superfamily reveals the unexplored diversity of pogo-like elements.Mob DNA. 2020 Jun 29;11:21. doi: 10.1186/s13100-020-00212-0. eCollection 2020. Mob DNA. 2020. PMID: 32612713 Free PMC article.
-
Modern thoughts on an ancyent marinere: function, evolution, regulation.Annu Rev Genet. 1997;31:337-58. doi: 10.1146/annurev.genet.31.1.337. Annu Rev Genet. 1997. PMID: 9442899 Review.
-
Assembly of the Tc1 and mariner transposition initiation complexes depends on the origins of their transposase DNA binding domains.Genetica. 2007 Jun;130(2):105-20. doi: 10.1007/s10709-006-0025-2. Epub 2006 Aug 16. Genetica. 2007. PMID: 16912840 Review.
Cited by
-
Different Families of Retrotransposons and DNA Transposons Are Actively Transcribed and May Have Transposed Recently in Physcomitrium (Physcomitrella) patens.Front Plant Sci. 2020 Aug 19;11:1274. doi: 10.3389/fpls.2020.01274. eCollection 2020. Front Plant Sci. 2020. PMID: 32973835 Free PMC article.
-
TRT, a Vertebrate and Protozoan Tc1-Like Transposon: Current Activity and Horizontal Transfer.Genome Biol Evol. 2016 Oct 5;8(9):2994-3005. doi: 10.1093/gbe/evw213. Genome Biol Evol. 2016. PMID: 27667131 Free PMC article.
-
Introduction of Plant Transposon Annotation for Beginners.Biology (Basel). 2023 Nov 26;12(12):1468. doi: 10.3390/biology12121468. Biology (Basel). 2023. PMID: 38132293 Free PMC article. Review.
-
An Analysis of IS630/Tc1/mariner Transposons in the Genome of a Pacific Oyster, Crassostrea gigas.J Mol Evol. 2018 Oct;86(8):566-580. doi: 10.1007/s00239-018-9868-2. Epub 2018 Oct 3. J Mol Evol. 2018. PMID: 30283979
-
Intruder (DD38E), a recently evolved sibling family of DD34E/Tc1 transposons in animals.Mob DNA. 2020 Dec 10;11(1):32. doi: 10.1186/s13100-020-00227-7. Mob DNA. 2020. PMID: 33303022 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

