Two separate modules of the conserved regulatory RNA AbcR1 address multiple target mRNAs in and outside of the translation initiation region
- PMID: 24921646
- PMCID: PMC4152367
- DOI: 10.4161/rna.29145
Two separate modules of the conserved regulatory RNA AbcR1 address multiple target mRNAs in and outside of the translation initiation region
Abstract
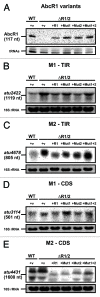
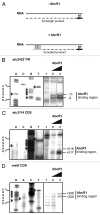
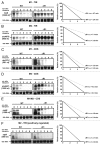
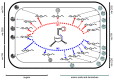
The small RNA AbcR1 regulates the expression of ABC transporters in the plant pathogen Agrobacterium tumefaciens, the plant symbiont Sinorhizobium meliloti, and the human pathogen Brucella abortus. A combination of proteomic and bioinformatic approaches suggested dozens of AbcR1 targets in A. tumefaciens. Several of these newly discovered targets are involved in the uptake of amino acids, their derivatives, and sugars. Among the latter is the periplasmic sugar-binding protein ChvE, a component of the virulence signal transduction system. We examined 16 targets and their interaction with AbcR1 in close detail. In addition to the previously described mRNA interaction site of AbcR1 (M1), the CopraRNA program predicted a second functional module (M2) as target-binding site. Both M1 and M2 contain single-stranded anti-SD motifs. Using mutated AbcR1 variants, we systematically tested by band shift experiments, which sRNA region is responsible for mRNA binding and gene regulation. On the target site, we find that AbcR1 interacts with some mRNAs in the translation initiation region and with others far into their coding sequence. Our data show that AbcR1 is a versatile master regulator of nutrient uptake systems in A. tumefaciens and related bacteria.
Keywords: ABC transporter; Agrobacterium; RNA-RNA interaction; alpha-proteobacteria; regulatory RNA; small RNA.
Figures




Similar articles
-
A 6-Nucleotide Regulatory Motif within the AbcR Small RNAs of Brucella abortus Mediates Host-Pathogen Interactions.mBio. 2017 Jun 6;8(3):e00473-17. doi: 10.1128/mBio.00473-17. mBio. 2017. PMID: 28588127 Free PMC article.
-
Independent activity of the homologous small regulatory RNAs AbcR1 and AbcR2 in the legume symbiont Sinorhizobium meliloti.PLoS One. 2013 Jul 15;8(7):e68147. doi: 10.1371/journal.pone.0068147. Print 2013. PLoS One. 2013. PMID: 23869210 Free PMC article.
-
Riboregulation in plant-associated α-proteobacteria.RNA Biol. 2014;11(5):550-62. doi: 10.4161/rna.29625. Epub 2014 Jul 8. RNA Biol. 2014. PMID: 25003187 Free PMC article. Review.
-
Genome-wide profiling of Hfq-binding RNAs uncovers extensive post-transcriptional rewiring of major stress response and symbiotic regulons in Sinorhizobium meliloti.RNA Biol. 2014;11(5):563-79. doi: 10.4161/rna.28239. Epub 2014 Feb 26. RNA Biol. 2014. PMID: 24786641 Free PMC article.
-
Small Noncoding RNAs in Agrobacterium tumefaciens.Curr Top Microbiol Immunol. 2018;418:195-213. doi: 10.1007/82_2018_84. Curr Top Microbiol Immunol. 2018. PMID: 29556823 Review.
Cited by
-
IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions.Nucleic Acids Res. 2017 Jul 3;45(W1):W435-W439. doi: 10.1093/nar/gkx279. Nucleic Acids Res. 2017. PMID: 28472523 Free PMC article.
-
Agrobacterium tumefaciens: A Bacterium Primed for Synthetic Biology.Biodes Res. 2020 May 26;2020:8189219. doi: 10.34133/2020/8189219. eCollection 2020. Biodes Res. 2020. PMID: 37849895 Free PMC article. Review.
-
Proteomics of Brucella.Proteomes. 2020 Apr 22;8(2):8. doi: 10.3390/proteomes8020008. Proteomes. 2020. PMID: 32331335 Free PMC article. Review.
-
The Small Non-coding RNA rli106 Contributes to the Environmental Adaptation and Pathogenicity of Listeria Monocytogenes.J Vet Res. 2023 Mar 17;67(1):67-77. doi: 10.2478/jvetres-2023-0013. eCollection 2023 Mar. J Vet Res. 2023. PMID: 37008770 Free PMC article.
-
An sRNA and Cold Shock Protein Homolog-Based Feedforward Loop Post-transcriptionally Controls Cell Cycle Master Regulator CtrA.Front Microbiol. 2018 Apr 24;9:763. doi: 10.3389/fmicb.2018.00763. eCollection 2018. Front Microbiol. 2018. PMID: 29740411 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
