Sorting cells for basal and induced autophagic flux by quantitative ratiometric flow cytometry
- PMID: 24915460
- PMCID: PMC4203556
- DOI: 10.4161/auto.29394
Sorting cells for basal and induced autophagic flux by quantitative ratiometric flow cytometry
Abstract
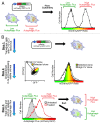
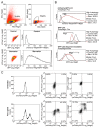
We detail here a protocol using tandem-tagged mCherry-EGFP-LC3 (C-G-LC3) to quantify autophagic flux in single cells by ratiometric flow cytometry and to isolate subpopulations of cells based on their relative levels of autophagic flux. This robust and sensitive method measures autophagic flux rather than autophagosome number and is an important addition to the autophagy researcher's array of tools for measuring autophagy. Two crucial steps in this protocol are i) generate cells constitutively expressing C-G-LC3 with low to medium fluorescence and low fluorescence variability, and ii) correctly set up gates and voltage/gain on a properly equipped flow cytometer. We have used this method to measure autophagic flux in a variety of cell types and experimental systems using many different autophagy stimuli. On a sorting flow cytometer, this technique can be used to isolate cells with different levels of basal autophagic flux, or cells with variable induction of flux in response to a given stimulus for further analysis or experimentation. We have also combined quantification of autophagic flux with methods to measure apoptosis and cell surface proteins, demonstrating the usefulness of this protocol in combination with other flow cytometry labels and markers.
Keywords: GFP-LC3; autophagic flux; autophagy; cell sorting; flow cytometry; quantification.
Figures
Similar articles
-
Monitoring autophagic flux by an improved tandem fluorescent-tagged LC3 (mTagRFP-mWasabi-LC3) reveals that high-dose rapamycin impairs autophagic flux in cancer cells.Autophagy. 2012 Aug;8(8):1215-26. doi: 10.4161/auto.20284. Epub 2012 May 31. Autophagy. 2012. PMID: 22647982
-
Flow cytometric analysis of autophagy in living mammalian cells.Methods Enzymol. 2009;452:131-41. doi: 10.1016/S0076-6879(08)03609-4. Methods Enzymol. 2009. PMID: 19200880
-
Establishment of a system for screening autophagic flux regulators using a modified fluorescent reporter and CRISPR/Cas9.Biochem Biophys Res Commun. 2019 Aug 27;516(3):686-692. doi: 10.1016/j.bbrc.2019.06.129. Epub 2019 Jun 26. Biochem Biophys Res Commun. 2019. PMID: 31253397
-
Quantifying autophagy: Measuring LC3 puncta and autolysosome formation in cells using multispectral imaging flow cytometry.Methods. 2017 Jan 1;112:147-156. doi: 10.1016/j.ymeth.2016.05.022. Epub 2016 Jun 1. Methods. 2017. PMID: 27263026 Review.
-
Autophagy: assays and artifacts.J Pathol. 2010 Jun;221(2):117-24. doi: 10.1002/path.2694. J Pathol. 2010. PMID: 20225337 Free PMC article. Review.
Cited by
-
Design, Synthesis, and Characterization of an Orally Active Dual-Specific ULK1/2 Autophagy Inhibitor that Synergizes with the PARP Inhibitor Olaparib for the Treatment of Triple-Negative Breast Cancer.J Med Chem. 2020 Dec 10;63(23):14609-14625. doi: 10.1021/acs.jmedchem.0c00873. Epub 2020 Nov 17. J Med Chem. 2020. PMID: 33200929 Free PMC article.
-
Autophagy Induction by HIV-Tat and Methamphetamine in Primary Midbrain Neuronal Cells of Tree Shrews via the mTOR Signaling and ATG5/ATG7 Pathway.Front Neurosci. 2018 Dec 6;12:921. doi: 10.3389/fnins.2018.00921. eCollection 2018. Front Neurosci. 2018. PMID: 30574066 Free PMC article.
-
Autophagy and Lysosomal Functionality in CMT2B Fibroblasts Carrying the RAB7K126R Mutation.Cells. 2022 Jan 31;11(3):496. doi: 10.3390/cells11030496. Cells. 2022. PMID: 35159308 Free PMC article.
-
The Dual Role of Autophagy in Crizotinib-Treated ALK+ ALCL: From the Lymphoma Cells Drug Resistance to Their Demise.Cells. 2021 Sep 23;10(10):2517. doi: 10.3390/cells10102517. Cells. 2021. PMID: 34685497 Free PMC article. Review.
-
Cancer Cells Upregulate NRF2 Signaling to Adapt to Autophagy Inhibition.Dev Cell. 2019 Sep 23;50(6):690-703.e6. doi: 10.1016/j.devcel.2019.07.010. Epub 2019 Aug 1. Dev Cell. 2019. PMID: 31378590 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
