Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine
- PMID: 24913268
- PMCID: PMC4086275
- DOI: 10.7554/eLife.03080
Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine
Abstract
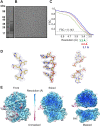
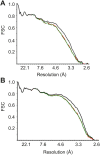
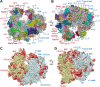
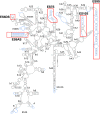
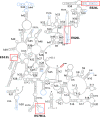
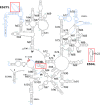
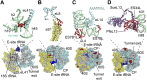
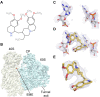
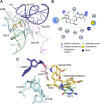
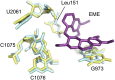
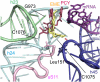
Malaria inflicts an enormous burden on global human health. The emergence of parasite resistance to front-line drugs has prompted a renewed focus on the repositioning of clinically approved drugs as potential anti-malarial therapies. Antibiotics that inhibit protein translation are promising candidates for repositioning. We have solved the cryo-EM structure of the cytoplasmic ribosome from the human malaria parasite, Plasmodium falciparum, in complex with emetine at 3.2 Å resolution. Emetine is an anti-protozoan drug used in the treatment of ameobiasis that also displays potent anti-malarial activity. Emetine interacts with the E-site of the ribosomal small subunit and shares a similar binding site with the antibiotic pactamycin, thereby delivering its therapeutic effect by blocking mRNA/tRNA translocation. As the first cryo-EM structure that visualizes an antibiotic bound to any ribosome at atomic resolution, this establishes cryo-EM as a powerful tool for screening and guiding the design of drugs that target parasite translation machinery.
Keywords: Plasmodium falciparum; biophysics; cryo-EM; drug development; malaria; ribosome; structural biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
Similar articles
-
Dynamical features of the Plasmodium falciparum ribosome during translation.Nucleic Acids Res. 2015 Dec 2;43(21):10515-24. doi: 10.1093/nar/gkv991. Epub 2015 Oct 1. Nucleic Acids Res. 2015. PMID: 26432834 Free PMC article.
-
Lead Optimization of Dehydroemetine for Repositioned Use in Malaria.Antimicrob Agents Chemother. 2020 Mar 24;64(4):e01444-19. doi: 10.1128/AAC.01444-19. Print 2020 Mar 24. Antimicrob Agents Chemother. 2020. PMID: 31964796 Free PMC article.
-
The Plasmodium falciparum cytoplasmic translation apparatus: a promising therapeutic target not yet exploited by clinically approved anti-malarials.Malar J. 2018 Dec 12;17(1):465. doi: 10.1186/s12936-018-2616-7. Malar J. 2018. PMID: 30541569 Free PMC article.
-
Ribosome dynamics: insights from atomic structure modeling into cryo-electron microscopy maps.Annu Rev Biophys Biomol Struct. 2006;35:299-317. doi: 10.1146/annurev.biophys.35.040405.101950. Annu Rev Biophys Biomol Struct. 2006. PMID: 16689638 Review.
-
The impact of recent improvements in cryo-electron microscopy technology on the understanding of bacterial ribosome assembly.Nucleic Acids Res. 2017 Feb 17;45(3):1027-1040. doi: 10.1093/nar/gkw1231. Nucleic Acids Res. 2017. PMID: 28180306 Free PMC article. Review.
Cited by
-
Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome.Proc Natl Acad Sci U S A. 2016 Jul 5;113(27):7527-32. doi: 10.1073/pnas.1604790113. Epub 2016 Jun 21. Proc Natl Acad Sci U S A. 2016. PMID: 27330110 Free PMC article.
-
2.8-Å Cryo-EM Structure of the Large Ribosomal Subunit from the Eukaryotic Parasite Leishmania.Cell Rep. 2016 Jul 12;16(2):288-294. doi: 10.1016/j.celrep.2016.06.014. Epub 2016 Jun 30. Cell Rep. 2016. PMID: 27373148 Free PMC article.
-
The mRNA-bound proteome of the human malaria parasite Plasmodium falciparum.Genome Biol. 2016 Jul 5;17(1):147. doi: 10.1186/s13059-016-1014-0. Genome Biol. 2016. PMID: 27381095 Free PMC article.
-
Mechanistic Insight into the Reactivation of BCAII Enzyme from Denatured and Molten Globule States by Eukaryotic Ribosomes and Domain V rRNAs.PLoS One. 2016 Apr 21;11(4):e0153928. doi: 10.1371/journal.pone.0153928. eCollection 2016. PLoS One. 2016. PMID: 27099964 Free PMC article.
-
Developments, applications, and prospects of cryo-electron microscopy.Protein Sci. 2020 Apr;29(4):872-882. doi: 10.1002/pro.3805. Epub 2019 Dec 26. Protein Sci. 2020. PMID: 31854478 Free PMC article. Review.
References
-
- Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, McAlear MA, Moore PB, Noller HF, Ortega J, Panse VG, Ramakrishnan V, Spahn CM, Steitz TA, Tchorzewski M, Tollervey D, Warren AJ, Williamson JR, Wilson D, Yonath A, Yusupov M. 2014. A new system for naming ribosomal proteins. Current Opinion in Structural Biology 24:165–169. doi: 10.1016/j.sbi.2014.01.002 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

