Interplay of interlocus gene conversion and crossover in segmental duplications under a neutral scenario
- PMID: 24906640
- PMCID: PMC4132178
- DOI: 10.1534/g3.114.012435
Interplay of interlocus gene conversion and crossover in segmental duplications under a neutral scenario
Abstract
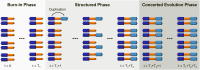
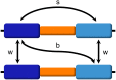
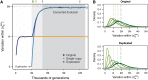
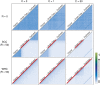
Interlocus gene conversion is a major evolutionary force that drives the concerted evolution of duplicated genomic regions. Theoretical models successfully have addressed the effects of interlocus gene conversion and the importance of crossover in the evolutionary fate of gene families and duplications but have not considered complex recombination scenarios, such as the presence of hotspots. To study the interplay between interlocus gene conversion and crossover, we have developed a forward-time simulator that allows the exploration of a wide range of interlocus gene conversion rates under different crossover models. Using it, we have analyzed patterns of nucleotide variation and linkage disequilibrium within and between duplicate regions, focusing on a neutral scenario with constant population size and validating our results with the existing theoretical models. We show that the interaction of gene conversion and crossover is nontrivial and that the location of crossover junctions is a fundamental determinant of levels of variation and linkage disequilibrium in duplicated regions. We also show that if crossover activity between duplications is strong enough, recurrent interlocus gene conversion events can break linkage disequilibrium within duplicates. Given the complex nature of interlocus gene conversion and crossover, we provide a framework to explore their interplay to help increase knowledge on molecular evolution within segmental duplications under more complex scenarios, such as demographic changes or natural selection.
Keywords: concerted evolution; forward simulations; increased variation; linkage disequilibrium; recombination hotspots.
Copyright © 2014 Hartasánchez et al.
Figures

Similar articles
-
Effect of Collapsed Duplications on Diversity Estimates: What to Expect.Genome Biol Evol. 2018 Nov 1;10(11):2899-2905. doi: 10.1093/gbe/evy223. Genome Biol Evol. 2018. PMID: 30364947 Free PMC article.
-
SeDuS: segmental duplication simulator.Bioinformatics. 2016 Jan 1;32(1):148-50. doi: 10.1093/bioinformatics/btv481. Epub 2015 Sep 10. Bioinformatics. 2016. PMID: 26358728
-
Signals of historical interlocus gene conversion in human segmental duplications.PLoS One. 2013 Oct 4;8(10):e75949. doi: 10.1371/journal.pone.0075949. eCollection 2013. PLoS One. 2013. PMID: 24124524 Free PMC article.
-
Role of selection in fixation of gene duplications.J Theor Biol. 2006 Mar 21;239(2):141-51. doi: 10.1016/j.jtbi.2005.08.033. Epub 2005 Oct 20. J Theor Biol. 2006. PMID: 16242725 Review.
-
Crossover Interference: Shedding Light on the Evolution of Recombination.Annu Rev Genet. 2019 Dec 3;53:19-44. doi: 10.1146/annurev-genet-040119-093957. Epub 2019 Aug 20. Annu Rev Genet. 2019. PMID: 31430178 Free PMC article. Review.
Cited by
-
Increased mutation and gene conversion within human segmental duplications.Nature. 2023 May;617(7960):325-334. doi: 10.1038/s41586-023-05895-y. Epub 2023 May 10. Nature. 2023. PMID: 37165237 Free PMC article.
-
Promoter evolution of mammalian gene duplicates.BMC Biol. 2023 Apr 13;21(1):80. doi: 10.1186/s12915-023-01590-6. BMC Biol. 2023. PMID: 37055747 Free PMC article.
-
Population Genetics of the Highly Polymorphic RPP8 Gene Family.Genes (Basel). 2019 Sep 8;10(9):691. doi: 10.3390/genes10090691. Genes (Basel). 2019. PMID: 31500388 Free PMC article.
-
A multi-omics data simulator for complex disease studies and its application to evaluate multi-omics data analysis methods for disease classification.Gigascience. 2019 May 1;8(5):giz045. doi: 10.1093/gigascience/giz045. Gigascience. 2019. PMID: 31029063 Free PMC article.
-
Effect of Collapsed Duplications on Diversity Estimates: What to Expect.Genome Biol Evol. 2018 Nov 1;10(11):2899-2905. doi: 10.1093/gbe/evy223. Genome Biol Evol. 2018. PMID: 30364947 Free PMC article.
References
-
- Bailey J. A., Eichler E. E., 2006. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat. Rev. Genet. 7: 552–564 - PubMed
-
- Bailey J. A., Gu Z., Clark R. A., Reinert K., Samonte R. V., et al. , 2002. Recent segmental duplications in the human genome. Science 297: 1003–1007 - PubMed
-
- Baltimore D., 1981. Gene conversion: some implications for immunoglobulin genes. Cell 24: 592–594 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
