Mechanism and function of oxidative reversal of DNA and RNA methylation
- PMID: 24905787
- PMCID: PMC4786441
- DOI: 10.1146/annurev-biochem-060713-035513
Mechanism and function of oxidative reversal of DNA and RNA methylation
Abstract
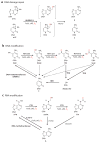
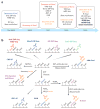
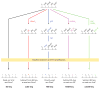
The importance of eukaryotic DNA methylation [5-methylcytosine (5mC)] in transcriptional regulation and development was first suggested almost 40 years ago. However, the molecular mechanism underlying the dynamic nature of this epigenetic mark was not understood until recently, following the discovery that the TET proteins, a family of AlkB-like Fe(II)/α-ketoglutarate-dependent dioxygenases, can oxidize 5mC to generate 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). Since then, several mechanisms that are responsible for processing oxidized 5mC derivatives to achieve DNA demethylation have emerged. Our biochemical understanding of the DNA demethylation process has prompted new investigations into the biological functions of DNA demethylation. Characterization of two additional AlkB family proteins, FTO and ALKBH5, showed that they possess demethylase activity toward N(6)-methyladenosine (m(6)A) in RNA, indicating that members of this subfamily of dioxygenases have a general function in demethylating nucleic acids. In this review, we discuss recent advances in this emerging field, focusing on the mechanism and function of TET-mediated DNA demethylation.
Keywords: 5-methylcytosine oxidation; DNA/RNA demethylation; Fe(II)/α-ketoglutarate-dependent dioxygenases; N6-methyladenosine; Tet.
Figures
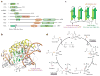


Similar articles
-
DNA repair enzymes ALKBH2, ALKBH3, and AlkB oxidize 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine in vitro.Nucleic Acids Res. 2019 Jun 20;47(11):5522-5529. doi: 10.1093/nar/gkz395. Nucleic Acids Res. 2019. PMID: 31114894 Free PMC article.
-
Enzymatic analysis of Tet proteins: key enzymes in the metabolism of DNA methylation.Methods Enzymol. 2012;512:93-105. doi: 10.1016/B978-0-12-391940-3.00005-6. Methods Enzymol. 2012. PMID: 22910204 Free PMC article.
-
Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA.Nature. 2014 Feb 20;506(7488):391-5. doi: 10.1038/nature12905. Epub 2013 Dec 25. Nature. 2014. PMID: 24390346 Free PMC article.
-
TET-mediated active DNA demethylation: mechanism, function and beyond.Nat Rev Genet. 2017 Sep;18(9):517-534. doi: 10.1038/nrg.2017.33. Epub 2017 May 30. Nat Rev Genet. 2017. PMID: 28555658 Review.
-
Enzymatic DNA oxidation: mechanisms and biological significance.BMB Rep. 2014 Nov;47(11):609-18. doi: 10.5483/bmbrep.2014.47.11.223. BMB Rep. 2014. PMID: 25341925 Free PMC article. Review.
Cited by
-
5-Hydroxymethylcytosines in circulating cell-free DNA reveal a diagnostic biomarker for glioma.Heliyon. 2022 Oct 12;8(10):e11022. doi: 10.1016/j.heliyon.2022.e11022. eCollection 2022 Oct. Heliyon. 2022. PMID: 36281400 Free PMC article.
-
A positive-feedback loop between HBx and ALKBH5 promotes hepatocellular carcinogenesis.BMC Cancer. 2021 Jun 10;21(1):686. doi: 10.1186/s12885-021-08449-5. BMC Cancer. 2021. PMID: 34112124 Free PMC article.
-
Molecular basis of the plant ROS1-mediated active DNA demethylation.Nat Plants. 2023 Feb;9(2):271-279. doi: 10.1038/s41477-022-01322-8. Epub 2023 Jan 9. Nat Plants. 2023. PMID: 36624257
-
Highly Selective 5-Formyluracil Labeling and Genome-wide Mapping Using (2-Benzimidazolyl)Acetonitrile Probe.iScience. 2018 Nov 30;9:423-432. doi: 10.1016/j.isci.2018.10.024. Epub 2018 Nov 2. iScience. 2018. PMID: 30466066 Free PMC article.
-
5-Hydroxymethylcytosine and ten-eleven translocation dioxygenases in head and neck carcinoma.J Cancer. 2019 Aug 28;10(21):5306-5314. doi: 10.7150/jca.34806. eCollection 2019. J Cancer. 2019. PMID: 31602281 Free PMC article.
References
-
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. - PubMed
-
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. - PubMed
-
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

