Structural and functional characterization of MERS coronavirus papain-like protease
- PMID: 24898546
- PMCID: PMC4051379
- DOI: 10.1186/1423-0127-21-54
Structural and functional characterization of MERS coronavirus papain-like protease
Abstract
Backgrounds: A new highly pathogenic human coronavirus (CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), has emerged in Jeddah and Saudi Arabia and quickly spread to some European countries since September 2012. Until 15 May 2014, it has infected at least 572 people with a fatality rate of about 30% globally. Studies to understand the virus and to develop antiviral drugs or therapy are necessary and urgent. In the present study, MERS-CoV papain-like protease (PLpro) is expressed, and its structural and functional consequences are elucidated.
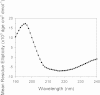
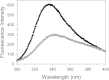
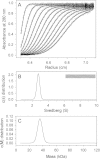
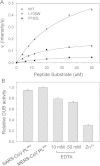
Results: Circular dichroism and Tyr/Trp fluorescence analyses indicated that the secondary and tertiary structure of MERS-CoV PLpro is well organized and folded. Analytical ultracentrifugation analyses demonstrated that MERS-CoV PLpro is a monomer in solution. The steady-state kinetic and deubiquitination activity assays indicated that MERS-CoV PLpro exhibits potent deubiquitination activity but lower proteolytic activity, compared with SARS-CoV PLpro. A natural mutation, Leu105, is the major reason for this difference.
Conclusions: Overall, MERS-CoV PLpro bound by an endogenous metal ion shows a folded structure and potent proteolytic and deubiquitination activity. These findings provide important insights into the structural and functional properties of coronaviral PLpro family, which is applicable to develop strategies inhibiting PLpro against highly pathogenic coronaviruses.
Figures

Similar articles
-
The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds.Antiviral Res. 2015 Mar;115:21-38. doi: 10.1016/j.antiviral.2014.12.015. Epub 2014 Dec 29. Antiviral Res. 2015. PMID: 25554382 Free PMC article. Review.
-
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV.ACS Chem Biol. 2015 Jun 19;10(6):1456-65. doi: 10.1021/cb500917m. Epub 2015 Mar 16. ACS Chem Biol. 2015. PMID: 25746232 Free PMC article.
-
Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease.J Gen Virol. 2014 Mar;95(Pt 3):614-626. doi: 10.1099/vir.0.059014-0. Epub 2013 Dec 20. J Gen Virol. 2014. PMID: 24362959
-
Structurally Guided Removal of DeISGylase Biochemical Activity from Papain-Like Protease Originating from Middle East Respiratory Syndrome Coronavirus.J Virol. 2017 Nov 14;91(23):e01067-17. doi: 10.1128/JVI.01067-17. Print 2017 Dec 1. J Virol. 2017. PMID: 28931677 Free PMC article.
-
Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease.Molecules. 2020 Aug 27;25(17):3920. doi: 10.3390/molecules25173920. Molecules. 2020. PMID: 32867349 Free PMC article. Review.
Cited by
-
A Computational Approach for Predicting Role of Human MicroRNAs in MERS-CoV Genome.Adv Bioinformatics. 2014;2014:967946. doi: 10.1155/2014/967946. Epub 2014 Dec 23. Adv Bioinformatics. 2014. PMID: 25610462 Free PMC article.
-
Middle East respiratory syndrome coronavirus: transmission, virology and therapeutic targeting to aid in outbreak control.Exp Mol Med. 2015 Aug 28;47(8):e181. doi: 10.1038/emm.2015.76. Exp Mol Med. 2015. PMID: 26315600 Free PMC article. Review.
-
Identification of novel allosteric sites of SARS-CoV-2 papain-like protease (PLpro) for the development of COVID-19 antivirals.J Biol Chem. 2024 Nov;300(11):107821. doi: 10.1016/j.jbc.2024.107821. Epub 2024 Sep 27. J Biol Chem. 2024. PMID: 39342997 Free PMC article.
-
COVID-19: fighting the invisible enemy with microRNAs.Expert Rev Anti Infect Ther. 2021 Feb;19(2):137-145. doi: 10.1080/14787210.2020.1812385. Epub 2020 Sep 16. Expert Rev Anti Infect Ther. 2021. PMID: 32814446 Free PMC article. Review.
-
Computed optical spectra of SARS-CoV-2 proteins.Chem Phys Lett. 2020 Nov;758:137935. doi: 10.1016/j.cplett.2020.137935. Epub 2020 Aug 29. Chem Phys Lett. 2020. PMID: 33518776 Free PMC article.
References
-
- Eckerle I, Muller MA, Kallies S, Gotthardt DN, Drosten C. In-vitro renal epithelial cell infection reveals a viral kidney tropism as a potential mechanism for acute renal failure during Middle East Respiratory Syndrome (MERS) Coronavirus infection. Virol J. 2013;10:359. doi: 10.1186/1743-422X-10-359. - DOI - PMC - PubMed
-
- Assiri A, McGeer A, Perl TM, Price CS, Al Rabeeah AA, Cummings DA, Alabdullatif ZN, Assad M, Almulhim A, Makhdoom H, Madani H, Alhakeem R, Al-Tawfiq JA, Cotten M, Watson SJ, Kellam P, Zumla AI, Memish ZA, Team KM-CI. Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013;369(5):407–416. doi: 10.1056/NEJMoa1306742. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

