One hundred million adenosine-to-inosine RNA editing sites: hearing through the noise
- PMID: 24889193
- PMCID: PMC4359916
- DOI: 10.1002/bies.201400055
One hundred million adenosine-to-inosine RNA editing sites: hearing through the noise
Abstract
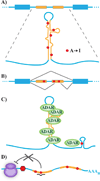
The most recent work toward compiling a comprehensive database of adenosine-to-inosine RNA editing events suggests that the potential for RNA editing is much more pervasive than previously thought; indeed, it is manifest in more than 100 million potential editing events located primarily within Alu repeat elements of the human transcriptome. Pairs of inverted Alu repeats are found in a substantial number of human genes, and when transcribed, they form long double-stranded RNA structures that serve as optimal substrates for RNA editing enzymes. A small subset of edited Alu elements has been shown to exhibit diverse functional roles in the regulation of alternative splicing, miRNA repression, and cis-regulation of distant RNA editing sites. The low level of editing for the remaining majority may be non-functional, yet their persistence in the primate genome provides enhanced genomic flexibility that may be required for adaptive evolution.
Keywords: ADAR; Alu; RNA editing; inosine.
© 2014 WILEY Periodicals, Inc.
Figures
Similar articles
-
A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes.Genome Res. 2014 Mar;24(3):365-76. doi: 10.1101/gr.164749.113. Epub 2013 Dec 17. Genome Res. 2014. PMID: 24347612 Free PMC article.
-
Alu elements shape the primate transcriptome by cis-regulation of RNA editing.Genome Biol. 2014 Feb 3;15(2):R28. doi: 10.1186/gb-2014-15-2-r28. Genome Biol. 2014. PMID: 24485196 Free PMC article.
-
ALU A-to-I RNA Editing: Millions of Sites and Many Open Questions.Methods Mol Biol. 2021;2181:149-162. doi: 10.1007/978-1-0716-0787-9_9. Methods Mol Biol. 2021. PMID: 32729079 Review.
-
Letter from the editor: Adenosine-to-inosine RNA editing in Alu repeats in the human genome.EMBO Rep. 2005 Sep;6(9):831-5. doi: 10.1038/sj.embor.7400507. EMBO Rep. 2005. PMID: 16138094 Free PMC article. Review.
-
Identification of widespread ultra-edited human RNAs.PLoS Genet. 2011 Oct;7(10):e1002317. doi: 10.1371/journal.pgen.1002317. Epub 2011 Oct 20. PLoS Genet. 2011. PMID: 22028664 Free PMC article.
Cited by
-
An Evolutionary Landscape of A-to-I RNA Editome across Metazoan Species.Genome Biol Evol. 2018 Feb 1;10(2):521-537. doi: 10.1093/gbe/evx277. Genome Biol Evol. 2018. PMID: 29294013 Free PMC article.
-
RNA editing landscape of adipose tissue in polycystic ovary syndrome provides insight into the obesity-related immune responses.Front Endocrinol (Lausanne). 2024 Jun 24;15:1379293. doi: 10.3389/fendo.2024.1379293. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 38978626 Free PMC article.
-
RNA-editing enzymes ADAR1 and ADAR2 coordinately regulate the editing and expression of Ctn RNA.FEBS Lett. 2017 Sep;591(18):2890-2904. doi: 10.1002/1873-3468.12795. Epub 2017 Aug 30. FEBS Lett. 2017. PMID: 28833069 Free PMC article.
-
ADAR2 enzymes: efficient site-specific RNA editors with gene therapy aspirations.RNA. 2022 Oct;28(10):1281-1297. doi: 10.1261/rna.079266.122. Epub 2022 Jul 21. RNA. 2022. PMID: 35863867 Free PMC article. Review.
-
Genome-wide analysis of consistently RNA edited sites in human blood reveals interactions with mRNA processing genes and suggests correlations with cell types and biological variables.BMC Genomics. 2018 Dec 27;19(1):963. doi: 10.1186/s12864-018-5364-8. BMC Genomics. 2018. PMID: 30587120 Free PMC article.
References
-
- Schmitz J. SINEs as driving forces in genome evolution. Genome dynamics. 2012;7:92–107. - PubMed
-
- Pullirsch D, Jantsch MF. Proteome diversification by adenosine to inosine RNA editing. RNA biology. 2010;7:205–212. - PubMed
-
- Wahlstedt H, Ohman M. Site-selective versus promiscuous A-to-I editing. Wiley interdisciplinary reviews RNA. 2011;2:761–771. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

