Transcriptomic changes during regeneration of the central nervous system in an echinoderm
- PMID: 24886271
- PMCID: PMC4229883
- DOI: 10.1186/1471-2164-15-357
Transcriptomic changes during regeneration of the central nervous system in an echinoderm
Abstract
Background: Echinoderms are emerging as important models in regenerative biology. Significant amount of data are available on cellular mechanisms of post-traumatic repair in these animals, whereas studies of gene expression are rare. In this study, we employ high-throughput sequencing to analyze the transcriptome of the normal and regenerating radial nerve cord (a homolog of the chordate neural tube), in the sea cucumber Holothuria glaberrima.
Results: Our de novo assembly yielded 70,173 contigs, of which 24,324 showed significant similarity to known protein-coding sequences. Expression profiling revealed large-scale changes in gene expression (4,023 and 3,257 up-regulated and down-regulated transcripts, respectively) associated with regeneration. Functional analysis of sets of differentially expressed genes suggested that among the most extensively over-represented pathways were those involved in the extracellular matrix (ECM) remodeling and ECM-cell interactions, indicating a key role of the ECM in regeneration. We also searched the sea cucumber transcriptome for homologs of factors known to be involved in acquisition and/or control of pluripotency. We identified eleven genes that were expressed both in the normal and regenerating tissues. Of these, only Myc was present at significantly higher levels in regeneration, whereas the expression of Bmi-1 was significantly reduced. We also sought to get insight into which transcription factors may operate at the top of the regulatory hierarchy to control gene expression in regeneration. Our analysis yielded eleven putative transcription factors, which constitute good candidates for further functional studies. The identified candidate transcription factors included not only known regeneration-related genes, but also factors not previously implicated as regulators of post-traumatic tissue regrowth. Functional annotation also suggested that one of the possible adaptations contributing to fast and efficient neural regeneration in echinoderms may be related to suppression of excitotoxicity.
Conclusions: Our transcriptomic analysis corroborates existing data on cellular mechanisms implicated in regeneration in sea cucumbers. More importantly, however, it also illuminates new aspects of echinoderm regeneration, which have been scarcely studied or overlooked altogether. The most significant outcome of the present work is that it lays out a roadmap for future studies of regulatory mechanisms by providing a list of key candidate genes for functional analysis.
Figures


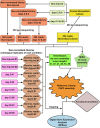
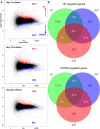
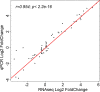

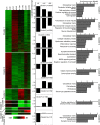
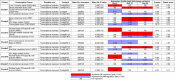


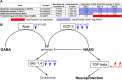
Similar articles
-
Transcriptomic analysis of sea cucumber (Holothuria leucospilota) coelomocytes revealed the echinoderm cytokine response during immune challenge.BMC Genomics. 2020 Apr 16;21(1):306. doi: 10.1186/s12864-020-6698-6. BMC Genomics. 2020. PMID: 32299355 Free PMC article.
-
Long Non-Coding RNAs (lncRNAs) of Sea Cucumber: Large-Scale Prediction, Expression Profiling, Non-Coding Network Construction, and lncRNA-microRNA-Gene Interaction Analysis of lncRNAs in Apostichopus japonicus and Holothuria glaberrima During LPS Challenge and Radial Organ Complex Regeneration.Mar Biotechnol (NY). 2016 Aug;18(4):485-99. doi: 10.1007/s10126-016-9711-y. Epub 2016 Jul 9. Mar Biotechnol (NY). 2016. PMID: 27392411
-
Ependymin, a gene involved in regeneration and neuroplasticity in vertebrates, is overexpressed during regeneration in the echinoderm Holothuria glaberrima.Gene. 2004 Jun 9;334:133-43. doi: 10.1016/j.gene.2004.03.023. Gene. 2004. PMID: 15256263
-
Holothurians as a Model System to Study Regeneration.Results Probl Cell Differ. 2018;65:255-283. doi: 10.1007/978-3-319-92486-1_13. Results Probl Cell Differ. 2018. PMID: 30083924 Review.
-
A roadmap for intestinal regeneration.Int J Dev Biol. 2021;65(4-5-6):427-437. doi: 10.1387/ijdb.200227dq. Int J Dev Biol. 2021. PMID: 32930367 Free PMC article. Review.
Cited by
-
Transcriptomic analysis reveals the early body wall regeneration mechanism of the sea cucumber Holothuria leucospilota after artificially induced transverse fission.BMC Genomics. 2023 Dec 12;24(1):766. doi: 10.1186/s12864-023-09808-1. BMC Genomics. 2023. PMID: 38087211 Free PMC article.
-
Transcriptomic analysis of sea cucumber (Holothuria leucospilota) coelomocytes revealed the echinoderm cytokine response during immune challenge.BMC Genomics. 2020 Apr 16;21(1):306. doi: 10.1186/s12864-020-6698-6. BMC Genomics. 2020. PMID: 32299355 Free PMC article.
-
Identification and localization of growth factor genes in the sea cucumber, Holothuria scabra.Heliyon. 2021 Nov 13;7(11):e08370. doi: 10.1016/j.heliyon.2021.e08370. eCollection 2021 Nov. Heliyon. 2021. PMID: 34825084 Free PMC article.
-
Analysis of sea star larval regeneration reveals conserved processes of whole-body regeneration across the metazoa.BMC Biol. 2019 Feb 22;17(1):16. doi: 10.1186/s12915-019-0633-9. BMC Biol. 2019. PMID: 30795750 Free PMC article.
-
Molecular characterization and gene expression patterns of retinoid receptors, in normal and regenerating tissues of the sea cucumber, Holothuria glaberrima.Gene. 2018 May 15;654:23-35. doi: 10.1016/j.gene.2018.01.102. Epub 2018 Feb 7. Gene. 2018. PMID: 29425825 Free PMC article.
References
-
- Mashanov VS, Zueva OR, García-Arrarás JE. Radial glial cells play a key role in echinoderm neural regeneration. BMC Biol. 2013;11:49. [ http://www.biomedcentral.com/1741-7007/11/49] - PMC - PubMed
-
- Mashanov VS, Zueva O, García-Arrarás JE. Mechanisms of Regeneration, Volume 108 of Current Topics in Developmental Biology. San Diego: Academic Press; 2014. Chapter seven - postembryonic organogenesis of the digestive tube: why does it occur in worms and sea cucumbers but fail in humans? pp. 185–216. [ http://www.sciencedirect.com/science/article/pii/B9780123914989000061] - PMC - PubMed
-
- Rojas-Cartagena C, Ortíz-Pineda P, Ramírez-Gómez F, Suárez-Castillo EC, Matos-Cruz V, Rodríguez C, Ortíz-Zuazaga H, García-Arrarás JE. Distinct profiles of expressed sequence tags during intestinal regeneration in the sea cucumber Holothuria glaberrima. Physiol Genomics. 2007;31(2):203–215. - PMC - PubMed
-
- Sun L, Chen M, Yang H, Wang T, Liu B, Shu C, Gardiner DM. Large scale gene expression profiling during intestine and body wall regeneration in the sea cucumber Apostichopus japonicus. Comp Biochem Physiol Part D Genomics Proteomics. 2011;6(2):195–205. - PubMed
-
- Heinzeller T, Welsch U. In: Brain evolution and cognition. Roth G, Wullimann M, editor. New York: Wiley-Spektrum; 2001. The echinoderm nervous system and its phylogenetic interpretation; pp. 41–75.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

