Multiple transcription factors directly regulate Hox gene lin-39 expression in ventral hypodermal cells of the C. elegans embryo and larva, including the hypodermal fate regulators LIN-26 and ELT-6
- PMID: 24885717
- PMCID: PMC4051164
- DOI: 10.1186/1471-213X-14-17
Multiple transcription factors directly regulate Hox gene lin-39 expression in ventral hypodermal cells of the C. elegans embryo and larva, including the hypodermal fate regulators LIN-26 and ELT-6
Abstract
Background: Hox genes encode master regulators of regional fate specification during early metazoan development. Much is known about the initiation and regulation of Hox gene expression in Drosophila and vertebrates, but less is known in the non-arthropod invertebrate model system, C. elegans. The C. elegans Hox gene lin-39 is required for correct fate specification in the midbody region, including the Vulval Precursor Cells (VPCs). To better understand lin-39 regulation and function, we aimed to identify transcription factors necessary for lin-39 expression in the VPCs, and in particular sought factors that initiate lin-39 expression in the embryo.
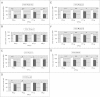
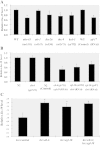
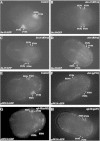
Results: We used the yeast one-hybrid (Y1H) method to screen for factors that bound to 13 fragments from the lin-39 region: twelve fragments contained sequences conserved between C. elegans and two other nematode species, while one fragment was known to drive reporter gene expression in the early embryo in cells that generate the VPCs. Sixteen transcription factors that bind to eight lin-39 genomic fragments were identified in yeast, and we characterized several factors by verifying their physical interactions in vitro, and showing that reduction of their function leads to alterations in lin-39 levels and lin-39::GFP reporter expression in vivo. Three factors, the orphan nuclear hormone receptor NHR-43, the hypodermal fate regulator LIN-26, and the GATA factor ELT-6 positively regulate lin-39 expression in the embryonic precursors to the VPCs. In particular, ELT-6 interacts with an enhancer that drives GFP expression in the early embryo, and the ELT-6 site we identified is necessary for proper embryonic expression. These three factors, along with the factors ZTF-17, BED-3 and TBX-9, also positively regulate lin-39 expression in the larval VPCs.
Conclusions: These results significantly expand the number of factors known to directly bind and regulate lin-39 expression, identify the first factors required for lin-39 expression in the embryo, and hint at a positive feedback mechanism involving GATA factors that maintains lin-39 expression in the vulval lineage. This work indicates that, as in other organisms, the regulation of Hox gene expression in C. elegans is complicated, redundant and robust.
Figures




Similar articles
-
Cell fates and fusion in the C. elegans vulval primordium are regulated by the EGL-18 and ELT-6 GATA factors -- apparent direct targets of the LIN-39 Hox protein.Development. 2002 Nov;129(22):5171-80. doi: 10.1242/dev.129.22.5171. Development. 2002. PMID: 12399309
-
The Paired-box protein PAX-3 regulates the choice between lateral and ventral epidermal cell fates in C. elegans.Dev Biol. 2016 Apr 15;412(2):191-207. doi: 10.1016/j.ydbio.2016.03.002. Epub 2016 Mar 4. Dev Biol. 2016. PMID: 26953187 Free PMC article.
-
ELT-3: A Caenorhabditis elegans GATA factor expressed in the embryonic epidermis during morphogenesis.Dev Biol. 1999 Apr 15;208(2):265-80. doi: 10.1006/dbio.1999.9202. Dev Biol. 1999. PMID: 10191044
-
Signal transduction and cell fate specification during Caenorhabditis elegans vulval development.Curr Opin Genet Dev. 1994 Aug;4(4):508-16. doi: 10.1016/0959-437x(94)90065-b. Curr Opin Genet Dev. 1994. PMID: 7950317 Review.
-
Maintenance of neuronal identity in C. elegans and beyond: Lessons from transcription and chromatin factors.Semin Cell Dev Biol. 2024 Feb 15;154(Pt A):35-47. doi: 10.1016/j.semcdb.2023.07.001. Epub 2023 Jul 11. Semin Cell Dev Biol. 2024. PMID: 37438210 Free PMC article. Review.
Cited by
-
Identification of Wnt Pathway Target Genes Regulating the Division and Differentiation of Larval Seam Cells and Vulval Precursor Cells in Caenorhabditis elegans.G3 (Bethesda). 2015 Jun 5;5(8):1551-66. doi: 10.1534/g3.115.017715. G3 (Bethesda). 2015. PMID: 26048561 Free PMC article.
-
The C. elegans embryonic fate specification factor EGL-18 (GATA) is reutilized downstream of Wnt signaling to maintain a population of larval progenitor cells.Worm. 2015 Jan 27;4(1):e996419. doi: 10.1080/23723556.2014.996419. eCollection 2015 Jan-Mar. Worm. 2015. PMID: 26430560 Free PMC article.
References
-
- Foronda D, de Navas LF, Garaulet DL, Sanchez-Herrero E. Function and specificity of Hox genes. Int J Dev Biol. 2009;53(8–10):1404–1419. - PubMed
-
- Deutsch J. In: Advance in Experimental Medicine and Biology, Volume 689. Deutsch J, editor. Austin, Texas USA: Landes Bioscience; 2010. Hox Genes: Studies from the 20th to the 21st Century. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

