Selection on synonymous codons in mammalian rhodopsins: a possible role in optimizing translational processes
- PMID: 24884412
- PMCID: PMC4021273
- DOI: 10.1186/1471-2148-14-96
Selection on synonymous codons in mammalian rhodopsins: a possible role in optimizing translational processes
Abstract
Background: Synonymous codon usage can affect many cellular processes, particularly those associated with translation such as polypeptide elongation and folding, mRNA degradation/stability, and splicing. Highly expressed genes are thought to experience stronger selection pressures on synonymous codons. This should result in codon usage bias even in species with relatively low effective population sizes, like mammals, where synonymous site selection is thought to be weak. Here we use phylogenetic codon-based likelihood models to explore patterns of codon usage bias in a dataset of 18 mammalian rhodopsin sequences, the protein mediating the first step in vision in the eye, and one of the most highly expressed genes in vertebrates. We use these patterns to infer selection pressures on key translational mechanisms including polypeptide elongation, protein folding, mRNA stability, and splicing.
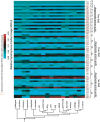
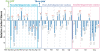
Results: Overall, patterns of selection in mammalian rhodopsin appear to be correlated with post-transcriptional and translational processes. We found significant evidence for selection at synonymous sites using phylogenetic mutation-selection likelihood models, with C-ending codons found to have the highest relative fitness, and to be significantly more abundant at conserved sites. In general, these codons corresponded with the most abundant tRNAs in mammals. We found significant differences in codon usage bias between rhodopsin loops versus helices, though there was no significant difference in mean synonymous substitution rate between these motifs. We also found a significantly higher proportion of GC-ending codons at paired sites in rhodopsin mRNA secondary structure, and significantly lower synonymous mutation rates in putative exonic splicing enhancer (ESE) regions than in non-ESE regions.
Conclusions: By focusing on a single highly expressed gene we both distinguish synonymous codon selection from mutational effects and analytically explore underlying functional mechanisms. Our results suggest that codon bias in mammalian rhodopsin arises from selection to optimally balance high overall translational speed, accuracy, and proper protein folding, especially in structurally complicated regions. Selection at synonymous sites may also be contributing to mRNA stability and splicing efficiency at exonic-splicing-enhancer (ESE) regions. Our results highlight the importance of investigating highly expressed genes in a broader phylogenetic context in order to better understand the evolution of synonymous substitutions.
Figures
Similar articles
-
Evidence for a trade-off between translational efficiency and splicing regulation in determining synonymous codon usage in Drosophila melanogaster.Mol Biol Evol. 2007 Dec;24(12):2755-62. doi: 10.1093/molbev/msm210. Epub 2007 Sep 28. Mol Biol Evol. 2007. PMID: 17905999
-
Molecular evolution of synonymous codon usage in Populus.BMC Evol Biol. 2008 Nov 4;8:307. doi: 10.1186/1471-2148-8-307. BMC Evol Biol. 2008. PMID: 18983655 Free PMC article.
-
Effect of exonic splicing regulation on synonymous codon usage in alternatively spliced exons of Dscam.BMC Evol Biol. 2009 Aug 27;9:214. doi: 10.1186/1471-2148-9-214. BMC Evol Biol. 2009. PMID: 19709440 Free PMC article.
-
[Synonymous Codon Usage-a Guide for Co-Translational Protein Folding in the Cell].Mol Biol (Mosk). 2019 Nov-Dec;53(6):883-898. doi: 10.1134/S0026898419060090. Mol Biol (Mosk). 2019. PMID: 31876270 Free PMC article. Review.
-
New insights into the factors affecting synonymous codon usage in human infecting Plasmodium species.Acta Trop. 2017 Dec;176:29-33. doi: 10.1016/j.actatropica.2017.07.025. Epub 2017 Jul 24. Acta Trop. 2017. PMID: 28751162 Review.
Cited by
-
Codon Usage Differences among Genes Expressed in Different Tissues of Drosophila melanogaster.Genome Biol Evol. 2019 Apr 1;11(4):1054-1065. doi: 10.1093/gbe/evz051. Genome Biol Evol. 2019. PMID: 30859203 Free PMC article.
-
Synonymous Site-to-Site Substitution Rate Variation Dramatically Inflates False Positive Rates of Selection Analyses: Ignore at Your Own Peril.Mol Biol Evol. 2020 Aug 1;37(8):2430-2439. doi: 10.1093/molbev/msaa037. Mol Biol Evol. 2020. PMID: 32068869 Free PMC article.
-
Decoding mechanisms by which silent codon changes influence protein biogenesis and function.Int J Biochem Cell Biol. 2015 Jul;64:58-74. doi: 10.1016/j.biocel.2015.03.011. Epub 2015 Mar 26. Int J Biochem Cell Biol. 2015. PMID: 25817479 Free PMC article. Review.
-
Rescue of mutant rhodopsin traffic by metformin-induced AMPK activation accelerates photoreceptor degeneration.Hum Mol Genet. 2017 Jan 15;26(2):305-319. doi: 10.1093/hmg/ddw387. Hum Mol Genet. 2017. PMID: 28065882 Free PMC article.
-
Next-generation development and application of codon model in evolution.Front Genet. 2023 Jan 27;14:1091575. doi: 10.3389/fgene.2023.1091575. eCollection 2023. Front Genet. 2023. PMID: 36777719 Free PMC article. Review.
References
-
- Li WH, Wu CI, Luo CC. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2(2):150–174. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

