A central role for the primary microRNA stem in guiding the position and efficiency of Drosha processing of a viral pri-miRNA
- PMID: 24854622
- PMCID: PMC4114686
- DOI: 10.1261/rna.044537.114
A central role for the primary microRNA stem in guiding the position and efficiency of Drosha processing of a viral pri-miRNA
Abstract
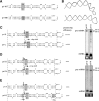
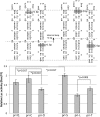
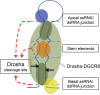
Processing of primary microRNA (pri-miRNA) stem-loops by the Drosha-DGCR8 complex is the initial step in miRNA maturation and crucial for miRNA function. Nonetheless, the underlying mechanism that determines the Drosha cleavage site of pri-miRNAs has remained unclear. Two prevalent but seemingly conflicting models propose that Drosha-DGCR8 anchors to and directs cleavage a fixed distance from either the basal single-stranded (ssRNA) or the terminal loop. However, recent studies suggest that the basal ssRNA and/or the terminal loop may influence the Drosha cleavage site dependent upon the sequence/structure of individual pri-miRNAs. Here, using a panel of closely related pri-miRNA variants, we further examine the role of pri-miRNA structures on Drosha cleavage site selection in cells. Our data reveal that both the basal ssRNA and terminal loop influence the Drosha cleavage site within three pri-miRNAs, the Simian Virus 40 (SV40) pri-miRNA, pri-miR-30a, and pri-miR-16. In addition to the flanking ssRNA regions, we show that an internal loop within the SV40 pri-miRNA stem strongly influences Drosha cleavage position and efficiency. We further demonstrate that the positions of the internal loop, basal ssRNA, and the terminal loop of the SV40 pri-miRNA cooperatively coordinate Drosha cleavage position and efficiency. Based on these observations, we propose that the pri-miRNA stem, defined by internal and flanking structural elements, guides the binding position of Drosha-DGCR8, which consequently determines the cleavage site. This study provides mechanistic insight into pri-miRNA processing in cells that has numerous biological implications and will assist in refining Drosha-dependent shRNA design.
Keywords: DGCR8; Drosha; RNAi; miRNA; pri-miRNA; shRNA.
© 2014 Burke et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures




Similar articles
-
Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex.Cell. 2006 Jun 2;125(5):887-901. doi: 10.1016/j.cell.2006.03.043. Cell. 2006. PMID: 16751099
-
Lower and upper stem-single-stranded RNA junctions together determine the Drosha cleavage site.Proc Natl Acad Sci U S A. 2013 Dec 17;110(51):20687-92. doi: 10.1073/pnas.1311639110. Epub 2013 Dec 2. Proc Natl Acad Sci U S A. 2013. PMID: 24297910 Free PMC article.
-
The core microprocessor component DiGeorge syndrome critical region 8 (DGCR8) is a nonspecific RNA-binding protein.J Biol Chem. 2013 Sep 13;288(37):26785-99. doi: 10.1074/jbc.M112.446880. Epub 2013 Jul 26. J Biol Chem. 2013. PMID: 23893406 Free PMC article.
-
The role of the precursor structure in the biogenesis of microRNA.Cell Mol Life Sci. 2011 Sep;68(17):2859-71. doi: 10.1007/s00018-011-0726-2. Epub 2011 May 24. Cell Mol Life Sci. 2011. PMID: 21607569 Free PMC article. Review.
-
Stimulation of pri-miR-18a Processing by hnRNP A1.Adv Exp Med Biol. 2011;700:28-35. doi: 10.1007/978-1-4419-7823-3_3. Adv Exp Med Biol. 2011. PMID: 21755470 Review.
Cited by
-
Identification of virus-encoded microRNAs in divergent Papillomaviruses.PLoS Pathog. 2018 Jul 26;14(7):e1007156. doi: 10.1371/journal.ppat.1007156. eCollection 2018 Jul. PLoS Pathog. 2018. PMID: 30048533 Free PMC article.
-
Structural Differences between Pri-miRNA Paralogs Promote Alternative Drosha Cleavage and Expand Target Repertoires.Cell Rep. 2019 Jan 8;26(2):447-459.e4. doi: 10.1016/j.celrep.2018.12.054. Cell Rep. 2019. PMID: 30625327 Free PMC article.
-
New epigenetic players in stroke pathogenesis: From non-coding RNAs to exosomal non-coding RNAs.Biomed Pharmacother. 2021 Aug;140:111753. doi: 10.1016/j.biopha.2021.111753. Epub 2021 May 25. Biomed Pharmacother. 2021. PMID: 34044272 Free PMC article. Review.
-
miRNAs derived from cancer-associated fibroblasts in colorectal cancer.Epigenomics. 2019 Nov;11(14):1627-1645. doi: 10.2217/epi-2019-0110. Epub 2019 Nov 8. Epigenomics. 2019. PMID: 31702390 Free PMC article. Review.
-
DUSP11 - An RNA phosphatase that regulates host and viral non-coding RNAs in mammalian cells.RNA Biol. 2017 Nov 2;14(11):1457-1465. doi: 10.1080/15476286.2017.1306169. Epub 2017 Apr 17. RNA Biol. 2017. PMID: 28296624 Free PMC article.
References
-
- Bartel DP 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 - PubMed
-
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. 2005. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 37: 766–770 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
