Coliphage HK022 Nun protein inhibits RNA polymerase translocation
- PMID: 24853501
- PMCID: PMC4060646
- DOI: 10.1073/pnas.1319740111
Coliphage HK022 Nun protein inhibits RNA polymerase translocation
Abstract
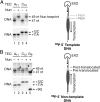
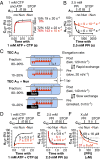
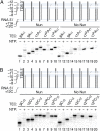
The Nun protein of coliphage HK022 arrests RNA polymerase (RNAP) in vivo and in vitro at pause sites distal to phage λ N-Utilization (nut) site RNA sequences. We tested the activity of Nun on ternary elongation complexes (TECs) assembled with templates lacking the λ nut sequence. We report that Nun stabilizes both translocation states of RNAP by restricting lateral movement of TEC along the DNA register. When Nun stabilized TEC in a pretranslocated register, immediately after NMP incorporation, it prevented binding of the next NTP and stimulated pyrophosphorolysis of the nascent transcript. In contrast, stabilization of TEC by Nun in a posttranslocated register allowed NTP binding and nucleotidyl transfer but inhibited pyrophosphorolysis and the next round of forward translocation. Nun binding to and action on the TEC requires a 9-bp RNA-DNA hybrid. We observed a Nun-dependent toe print upstream to the TEC. In addition, mutations in the RNAP β' subunit near the upstream end of the transcription bubble suppress Nun binding and arrest. These results suggest that Nun interacts with RNAP near the 5' edge of the RNA-DNA hybrid. By stabilizing translocation states through restriction of TEC lateral mobility, Nun represents a novel class of transcription arrest factors.
Keywords: Bacteriophage HK022; Bacteriophage Lambda; Exclusion; Transcription Elongation; Transcription Termination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


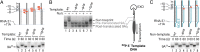
Similar articles
-
Structural basis of transcription arrest by coliphage HK022 Nun in an Escherichia coli RNA polymerase elongation complex.Elife. 2017 Mar 20;6:e25478. doi: 10.7554/eLife.25478. Elife. 2017. PMID: 28318486 Free PMC article.
-
Probing the structure of Nun transcription arrest factor bound to RNA polymerase.Proc Natl Acad Sci U S A. 2016 Aug 2;113(31):8693-8. doi: 10.1073/pnas.1601056113. Epub 2016 Jul 19. Proc Natl Acad Sci U S A. 2016. PMID: 27436904 Free PMC article.
-
The Nun protein of bacteriophage HK022 inhibits translocation of Escherichia coli RNA polymerase without abolishing its catalytic activities.Genes Dev. 1997 Oct 15;11(20):2670-8. doi: 10.1101/gad.11.20.2670. Genes Dev. 1997. PMID: 9334329 Free PMC article.
-
Structure and function in promoter escape by T7 RNA polymerase.Prog Nucleic Acid Res Mol Biol. 2005;80:323-47. doi: 10.1016/S0079-6603(05)80008-X. Prog Nucleic Acid Res Mol Biol. 2005. PMID: 16164978 Review. No abstract available.
-
Antitermination by bacteriophage lambda Q protein.Cold Spring Harb Symp Quant Biol. 1998;63:319-25. doi: 10.1101/sqb.1998.63.319. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384296 Review. No abstract available.
Cited by
-
Ancient Transcription Factors in the News.mBio. 2019 Feb 26;10(1):e01547-18. doi: 10.1128/mBio.01547-18. mBio. 2019. PMID: 30808693 Free PMC article. Review.
-
Transcription of Bacterial Chromatin.J Mol Biol. 2019 Sep 20;431(20):4040-4066. doi: 10.1016/j.jmb.2019.05.041. Epub 2019 May 31. J Mol Biol. 2019. PMID: 31153903 Free PMC article. Review.
-
HK022 Nun Requires Arginine-Rich Motif Residues Distinct from λ N.J Bacteriol. 2015 Nov;197(22):3573-82. doi: 10.1128/JB.00466-15. Epub 2015 Sep 8. J Bacteriol. 2015. PMID: 26350130 Free PMC article.
-
Bacteriophage HK022 Nun protein arrests transcription by blocking lateral mobility of RNA polymerase during transcription elongation.Bacteriophage. 2014 Jul 30;4:e32187. doi: 10.4161/bact.32187. eCollection 2014. Bacteriophage. 2014. PMID: 25105061 Free PMC article.
-
Native Mass Spectrometry-Based Screening for Optimal Sample Preparation in Single-Particle Cryo-EM.Structure. 2021 Feb 4;29(2):186-195.e6. doi: 10.1016/j.str.2020.11.001. Epub 2020 Nov 19. Structure. 2021. PMID: 33217329 Free PMC article.
References
-
- Bustamante C, Bryant Z, Smith SB. Ten years of tension: Single-molecule DNA mechanics. Nature. 2003;421(6921):423–427. - PubMed
-
- Galburt EA, et al. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446(7137):820–823. - PubMed
-
- Kireeva M, Kashlev M, Burton ZF. Translocation by multi-subunit RNA polymerases. Biochim Biophys Acta. 2010;1799(5-6):389–401. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

