Holocarboxylase synthetase interacts physically with nuclear receptor co-repressor, histone deacetylase 1 and a novel splicing variant of histone deacetylase 1 to repress repeats
- PMID: 24840043
- PMCID: PMC4743672
- DOI: 10.1042/BJ20131208
Holocarboxylase synthetase interacts physically with nuclear receptor co-repressor, histone deacetylase 1 and a novel splicing variant of histone deacetylase 1 to repress repeats
Abstract
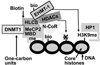
HLCS (holocarboxylase synthetase) is a nuclear protein that catalyses the binding of biotin to distinct lysine residues in chromatin proteins. HLCS-dependent epigenetic marks are over-represented in repressed genomic loci, particularly in repeats. Evidence is mounting that HLCS is a member of a multi-protein gene repression complex, which determines its localization in chromatin. In the present study we tested the hypothesis that HLCS interacts physically with N-CoR (nuclear receptor co-repressor) and HDAC1 (histone deacetylase 1), thereby contributing toward the removal of H3K9ac (Lys⁹-acetylated histone H3) gene activation marks and the repression of repeats. Physical interactions between HLCS and N-CoR, HDAC1 and a novel splicing variant of HDAC1 were confirmed by co-immunoprecipitation, limited proteolysis and split luciferase complementation assays. When HLCS was overexpressed, the abundance of H3K9ac marks decreased by 50% and 68% in LTRs (long terminal repeats) 15 and 22 respectively in HEK (human embryonic kidney)-293 cells compared with the controls. This loss of H3K9ac marks was linked with an 83% decrease in mRNA coding for LTRs. Similar patterns were seen in pericentromeric alpha satellite repeats in chromosomes 1 and 4. We conclude that interactions of HLCS with N-CoR and HDACs contribute towards the transcriptional repression of repeats, presumably increasing genome stability.
Figures





Similar articles
-
Holocarboxylase synthetase synergizes with methyl CpG binding protein 2 and DNA methyltransferase 1 in the transcriptional repression of long-terminal repeats.Epigenetics. 2013 May;8(5):504-11. doi: 10.4161/epi.24449. Epub 2013 Apr 27. Epigenetics. 2013. PMID: 23624957 Free PMC article.
-
Holocarboxylase synthetase interacts physically with euchromatic histone-lysine N-methyltransferase, linking histone biotinylation with methylation events.J Nutr Biochem. 2013 Aug;24(8):1446-52. doi: 10.1016/j.jnutbio.2012.12.003. Epub 2013 Jan 20. J Nutr Biochem. 2013. PMID: 23337344 Free PMC article.
-
Holocarboxylase synthetase acts as a biotin-independent transcriptional repressor interacting with HDAC1, HDAC2 and HDAC7.Mol Genet Metab. 2014 Mar;111(3):321-330. doi: 10.1016/j.ymgme.2013.10.016. Epub 2013 Nov 2. Mol Genet Metab. 2014. PMID: 24239178
-
A novel, enigmatic histone modification: biotinylation of histones by holocarboxylase synthetase.Nutr Rev. 2008 Dec;66(12):721-5. doi: 10.1111/j.1753-4887.2008.00127.x. Nutr Rev. 2008. PMID: 19019041 Review.
-
The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts.Biochem Soc Trans. 2013 Jun;41(3):741-9. doi: 10.1042/BST20130010. Biochem Soc Trans. 2013. PMID: 23697933 Review.
Cited by
-
Molecular Mechanisms of Biotin in Modulating Inflammatory Diseases.Nutrients. 2024 Jul 27;16(15):2444. doi: 10.3390/nu16152444. Nutrients. 2024. PMID: 39125325 Free PMC article. Review.
-
Biotin attenuates heat shock factor 4b transcriptional activity by lysine 444 biotinylation.Biochem Biophys Rep. 2022 Feb 5;30:101227. doi: 10.1016/j.bbrep.2022.101227. eCollection 2022 Jul. Biochem Biophys Rep. 2022. PMID: 35198740 Free PMC article.
-
Resveratrol compounds inhibit human holocarboxylase synthetase and cause a lean phenotype in Drosophila melanogaster.J Nutr Biochem. 2015 Nov;26(11):1379-84. doi: 10.1016/j.jnutbio.2015.07.004. Epub 2015 Jul 26. J Nutr Biochem. 2015. PMID: 26303405 Free PMC article.
-
Holocarboxylase synthetase knockout is embryonic lethal in mice.PLoS One. 2022 Apr 6;17(4):e0265539. doi: 10.1371/journal.pone.0265539. eCollection 2022. PLoS One. 2022. PMID: 35385533 Free PMC article.
-
β-Keto and β-hydroxyphosphonate analogs of biotin-5'-AMP are inhibitors of holocarboxylase synthetase.Bioorg Med Chem Lett. 2014 Dec 15;24(24):5568-5571. doi: 10.1016/j.bmcl.2014.11.010. Epub 2014 Nov 7. Bioorg Med Chem Lett. 2014. PMID: 25466176 Free PMC article.
References
-
- Suzuki Y, Aoki Y, Ishida Y, Chiba Y, Iwamatsu A, Kishino T, Niikawa N, Matsubara Y, Narisawa K. Isolation and characterization of mutations in the human holocarboxylase synthetase cDNA. Nat. Genet. 1994;8:122–128. - PubMed
-
- Zempleni J, Wijeratne SSK, Kuroishi T. Biotin. In: Erdman JW Jr, Macdonald I, Zeisel SH, editors. Present Knowledge in Nutrition. Washington: International Life Sciences Institute; 2012. pp. 587–609.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

