Papillomavirus genomes associate with BRD4 to replicate at fragile sites in the host genome
- PMID: 24832099
- PMCID: PMC4022725
- DOI: 10.1371/journal.ppat.1004117
Papillomavirus genomes associate with BRD4 to replicate at fragile sites in the host genome
Abstract
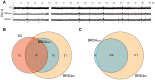
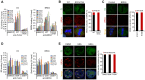
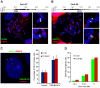
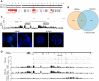
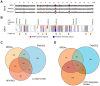
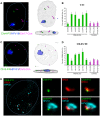
It has long been recognized that oncogenic viruses often integrate close to common fragile sites. The papillomavirus E2 protein, in complex with BRD4, tethers the viral genome to host chromatin to ensure persistent replication. Here, we map these targets to a number of large regions of the human genome and name them Persistent E2 and BRD4-Broad Localized Enrichments of Chromatin or PEB-BLOCs. PEB-BLOCs frequently contain deletions, have increased rates of asynchronous DNA replication, and are associated with many known common fragile sites. Cell specific fragile sites were mapped in human C-33 cervical cells by FANCD2 ChIP-chip, confirming the association with PEB-BLOCs. HPV-infected cells amplify viral DNA in nuclear replication foci and we show that these form adjacent to PEB-BLOCs. We propose that HPV replication, which hijacks host DNA damage responses, occurs adjacent to highly susceptible fragile sites, greatly increasing the chances of integration here, as is found in HPV-associated cancers.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Involvement of Brd4 in different steps of the papillomavirus life cycle.Virus Res. 2017 Mar 2;231:76-82. doi: 10.1016/j.virusres.2016.12.006. Epub 2016 Dec 10. Virus Res. 2017. PMID: 27965149 Free PMC article. Review.
-
Brd4 is displaced from HPV replication factories as they expand and amplify viral DNA.PLoS Pathog. 2013;9(11):e1003777. doi: 10.1371/journal.ppat.1003777. Epub 2013 Nov 21. PLoS Pathog. 2013. PMID: 24278023 Free PMC article.
-
Evidence supporting a role for TopBP1 and Brd4 in the initiation but not continuation of human papillomavirus 16 E1/E2-mediated DNA replication.J Virol. 2015 May;89(9):4980-91. doi: 10.1128/JVI.00335-15. Epub 2015 Feb 18. J Virol. 2015. PMID: 25694599 Free PMC article.
-
The human papillomavirus type 8 E2 tethering protein targets the ribosomal DNA loci of host mitotic chromosomes.J Virol. 2009 Jan;83(2):640-50. doi: 10.1128/JVI.01936-08. Epub 2008 Nov 12. J Virol. 2009. PMID: 19004936 Free PMC article.
-
Playing with fire: consequences of human papillomavirus DNA replication adjacent to genetically unstable regions of host chromatin.Curr Opin Virol. 2017 Oct;26:63-68. doi: 10.1016/j.coviro.2017.07.015. Epub 2017 Aug 3. Curr Opin Virol. 2017. PMID: 28779692 Review.
Cited by
-
Bioinformatics Analysis of Human Papillomavirus 16 Integration in Cervical Cancer: Changes in MAGI-1 Expression in Premalignant Lesions and Invasive Carcinoma.Cancers (Basel). 2024 Jun 14;16(12):2225. doi: 10.3390/cancers16122225. Cancers (Basel). 2024. PMID: 38927930 Free PMC article.
-
The NS1 protein of the parvovirus MVM Aids in the localization of the viral genome to cellular sites of DNA damage.PLoS Pathog. 2020 Oct 16;16(10):e1009002. doi: 10.1371/journal.ppat.1009002. eCollection 2020 Oct. PLoS Pathog. 2020. PMID: 33064772 Free PMC article.
-
An Epigenetic Compound Library Screen Identifies BET Inhibitors That Promote HSV-1 and -2 Replication by Bridging P-TEFb to Viral Gene Promoters through BRD4.PLoS Pathog. 2016 Oct 20;12(10):e1005950. doi: 10.1371/journal.ppat.1005950. eCollection 2016 Oct. PLoS Pathog. 2016. PMID: 27764245 Free PMC article.
-
Persistent Human Papillomavirus Infection.Viruses. 2021 Feb 20;13(2):321. doi: 10.3390/v13020321. Viruses. 2021. PMID: 33672465 Free PMC article. Review.
-
Involvement of Brd4 in different steps of the papillomavirus life cycle.Virus Res. 2017 Mar 2;231:76-82. doi: 10.1016/j.virusres.2016.12.006. Epub 2016 Dec 10. Virus Res. 2017. PMID: 27965149 Free PMC article. Review.
References
-
- You J, Croyle JL, Nishimura A, Ozato K, Howley PM (2004) Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117: 349–360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

