Hox gene regulation in the central nervous system of Drosophila
- PMID: 24795565
- PMCID: PMC4005941
- DOI: 10.3389/fncel.2014.00096
Hox gene regulation in the central nervous system of Drosophila
Abstract
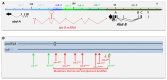
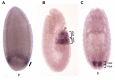
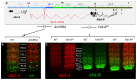
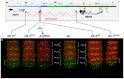
Hox genes specify the structures that form along the anteroposterior (AP) axis of bilateria. Within the genome, they often form clusters where, remarkably enough, their position within the clusters reflects the relative positions of the structures they specify along the AP axis. This correspondence between genomic organization and gene expression pattern has been conserved through evolution and provides a unique opportunity to study how chromosomal context affects gene regulation. In Drosophila, a general rule, often called "posterior dominance," states that Hox genes specifying more posterior structures repress the expression of more anterior Hox genes. This rule explains the apparent spatial complementarity of Hox gene expression patterns in Drosophila. Here we review a noticeable exception to this rule where the more-posteriorly expressed Abd-B Hox gene fails to repress the more-anterior abd-A gene in cells of the central nervous system (CNS). While Abd-B is required to repress ectopic expression of abd-A in the posterior epidermis, abd-A repression in the posterior CNS is accomplished by a different mechanism that involves a large 92 kb long non-coding RNA (lncRNA) encoded by the intergenic region separating abd-A and Abd-B (the iab8ncRNA). Dissection of this lncRNA revealed that abd-A is repressed by the lncRNA using two redundant mechanisms. The first mechanism is mediated by a microRNA (mir-iab-8) encoded by intronic sequence within the large iab8-ncRNA. Meanwhile, the second mechanism seems to involve transcriptional interference by the long iab-8 ncRNA on the abd-A promoter. Recent work demonstrating CNS-specific regulation of genes by ncRNAs in Drosophila, seem to highlight a potential role for the iab-8-ncRNA in the evolution of the Drosophila Hox complexes.
Keywords: Hox genes; abd-A; bithorax-complex; miRNA; ncRNA.
Figures

Similar articles
-
Following the intracellular localization of the iab-8ncRNA of the bithorax complex using the MS2-MCP-GFP system.Mech Dev. 2015 Nov;138 Pt 2:133-140. doi: 10.1016/j.mod.2015.08.004. Epub 2015 Aug 12. Mech Dev. 2015. PMID: 26277563
-
abd-A regulation by the iab-8 noncoding RNA.PLoS Genet. 2012;8(5):e1002720. doi: 10.1371/journal.pgen.1002720. Epub 2012 May 24. PLoS Genet. 2012. PMID: 22654672 Free PMC article.
-
Repression of the Hox gene abd-A by ELAV-mediated Transcriptional Interference.PLoS Genet. 2021 Nov 15;17(11):e1009843. doi: 10.1371/journal.pgen.1009843. eCollection 2021 Nov. PLoS Genet. 2021. PMID: 34780465 Free PMC article.
-
The bithorax complex of Drosophila an exceptional Hox cluster.Curr Top Dev Biol. 2009;88:1-33. doi: 10.1016/S0070-2153(09)88001-0. Curr Top Dev Biol. 2009. PMID: 19651300 Review.
-
A double-edged sword to force posterior dominance of Hox genes.Bioessays. 2008 Nov;30(11-12):1058-61. doi: 10.1002/bies.20847. Bioessays. 2008. PMID: 18937351 Review.
Cited by
-
Mechanisms of Long Non-Coding RNAs in the Assembly and Plasticity of Neural Circuitry.Front Neural Circuits. 2017 Oct 23;11:76. doi: 10.3389/fncir.2017.00076. eCollection 2017. Front Neural Circuits. 2017. PMID: 29109677 Free PMC article. Review.
-
The physiological function of long-noncoding RNAs.Noncoding RNA Res. 2020 Dec;5(4):178-184. doi: 10.1016/j.ncrna.2020.09.003. Epub 2020 Sep 17. Noncoding RNA Res. 2020. PMID: 32959025 Free PMC article. Review.
-
Role of abd-A and Abd-B in development of abdominal epithelia breaks posterior prevalence rule.PLoS Genet. 2014 Oct 23;10(10):e1004717. doi: 10.1371/journal.pgen.1004717. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25340649 Free PMC article.
-
Homeotic Genes: Clustering, Modularity, and Diversity.Front Cell Dev Biol. 2021 Aug 11;9:718308. doi: 10.3389/fcell.2021.718308. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34458272 Free PMC article. Review.
-
scRNA-seq data from the larval Drosophila ventral cord provides a resource for studying motor systems function and development.Dev Cell. 2024 May 6;59(9):1210-1230.e9. doi: 10.1016/j.devcel.2024.03.016. Epub 2024 Apr 2. Dev Cell. 2024. PMID: 38569548
References
-
- Barges S., Mihaly J., Galloni M., Hagstrom K., Müller M., Shanower G., et al. (2000). The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127 779–790 - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

