An integrative analysis of colon cancer identifies an essential function for PRPF6 in tumor growth
- PMID: 24788092
- PMCID: PMC4035536
- DOI: 10.1101/gad.237206.113
An integrative analysis of colon cancer identifies an essential function for PRPF6 in tumor growth
Abstract
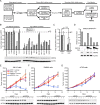
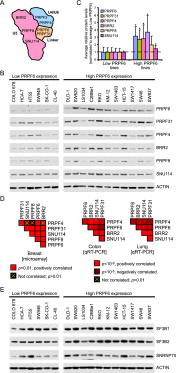
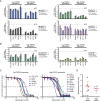
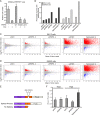
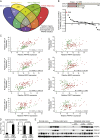
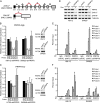
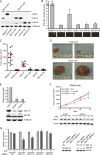
The spliceosome machinery is composed of multimeric protein complexes that generate a diverse repertoire of mRNA through coordinated splicing of heteronuclear RNAs. While somatic mutations in spliceosome components have been discovered in several cancer types, the molecular bases and consequences of spliceosome aberrations in cancer are poorly understood. Here we report for the first time that PRPF6, a member of the tri-snRNP (small ribonucleoprotein) spliceosome complex, drives cancer proliferation by preferential splicing of genes associated with growth regulation. Inhibition of PRPF6 and other tri-snRNP complex proteins, but not other snRNP spliceosome complexes, selectively abrogated growth in cancer cells with high tri-snRNP levels. High-resolution transcriptome analyses revealed that reduced PRPF6 alters the constitutive and alternative splicing of a discrete number of genes, including an oncogenic isoform of the ZAK kinase. These findings implicate an essential role for PRPF6 in cancer via splicing of distinct growth-related gene products.
Keywords: PRPF6; RNAi; colon cancer; spliceosome; tri-snRNP.
© 2014 Adler et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
Comment in
-
Alternative splicing: aberrant splicing promotes colon tumour growth.Nat Rev Cancer. 2014 Jun;14(6):382-3. doi: 10.1038/nrc3753. Nat Rev Cancer. 2014. PMID: 24854075 No abstract available.
Similar articles
-
A missense mutation in PRPF6 causes impairment of pre-mRNA splicing and autosomal-dominant retinitis pigmentosa.Am J Hum Genet. 2011 May 13;88(5):643-9. doi: 10.1016/j.ajhg.2011.04.008. Epub 2011 May 5. Am J Hum Genet. 2011. PMID: 21549338 Free PMC article.
-
SANS (USH1G) regulates pre-mRNA splicing by mediating the intra-nuclear transfer of tri-snRNP complexes.Nucleic Acids Res. 2021 Jun 4;49(10):5845-5866. doi: 10.1093/nar/gkab386. Nucleic Acids Res. 2021. PMID: 34023904 Free PMC article.
-
SMNrp is an essential pre-mRNA splicing factor required for the formation of the mature spliceosome.EMBO J. 2001 May 1;20(9):2304-14. doi: 10.1093/emboj/20.9.2304. EMBO J. 2001. PMID: 11331595 Free PMC article.
-
The Role of the U5 snRNP in Genetic Disorders and Cancer.Front Genet. 2021 Jan 28;12:636620. doi: 10.3389/fgene.2021.636620. eCollection 2021. Front Genet. 2021. PMID: 33584830 Free PMC article. Review.
-
How Is Precursor Messenger RNA Spliced by the Spliceosome?Annu Rev Biochem. 2020 Jun 20;89:333-358. doi: 10.1146/annurev-biochem-013118-111024. Epub 2019 Dec 9. Annu Rev Biochem. 2020. PMID: 31815536 Review.
Cited by
-
Hereditary retinoblastoma iPSC model reveals aberrant spliceosome function driving bone malignancies.Proc Natl Acad Sci U S A. 2022 Apr 19;119(16):e2117857119. doi: 10.1073/pnas.2117857119. Epub 2022 Apr 11. Proc Natl Acad Sci U S A. 2022. PMID: 35412907 Free PMC article.
-
Probing Isoform Switching Events in Various Cancer Types: Lessons From Pan-Cancer Studies.Front Mol Biosci. 2021 Nov 23;8:726902. doi: 10.3389/fmolb.2021.726902. eCollection 2021. Front Mol Biosci. 2021. PMID: 34888349 Free PMC article. Review.
-
Comparative proteogenomics profiling of non-small and small lung carcinoma cell lines using mass spectrometry.PeerJ. 2020 Apr 23;8:e8779. doi: 10.7717/peerj.8779. eCollection 2020. PeerJ. 2020. PMID: 32351780 Free PMC article.
-
Activation of the Classical Mitogen-Activated Protein Kinases Is Part of the Shiga Toxin-Induced Ribotoxic Stress Response and May Contribute to Shiga Toxin-Induced Inflammation.Infect Immun. 2015 Oct 19;84(1):138-48. doi: 10.1128/IAI.00977-15. Print 2016 Jan. Infect Immun. 2015. PMID: 26483408 Free PMC article.
-
The spliceosome is a therapeutic vulnerability in MYC-driven cancer.Nature. 2015 Sep 17;525(7569):384-8. doi: 10.1038/nature14985. Epub 2015 Sep 2. Nature. 2015. PMID: 26331541 Free PMC article.
References
-
- Adler AS, McCleland ML, Truong T, Lau S, Modrusan Z, Soukup TM, Roose-Girma M, Blackwood EM, Firestein R 2012. CDK8 maintains tumor dedifferentiation and embryonic stem cell pluripotency. Cancer Res 72: 2129–2139 - PubMed
-
- Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y, et al. 1989. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 244: 217–221 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
