Structural and functional insights to ubiquitin-like protein conjugation
- PMID: 24773014
- PMCID: PMC4118471
- DOI: 10.1146/annurev-biophys-051013-022958
Structural and functional insights to ubiquitin-like protein conjugation
Abstract
Attachment of ubiquitin (Ub) and ubiquitin-like proteins (Ubls) to cellular proteins regulates numerous cellular processes including transcription, the cell cycle, stress responses, DNA repair, apoptosis, immune responses, and autophagy, to name a few. The mechanistically parallel but functionally distinct conjugation pathways typically require the concerted activities of three types of protein: E1 Ubl-activating enzymes, E2 Ubl carrier proteins, and E3 Ubl ligases. E1 enzymes initiate pathway specificity for each cascade by recognizing and activating cognate Ubls, followed by catalyzing Ubl transfer to cognate E2 protein(s). Under certain circumstances, the E2 Ubl complex can direct ligation to the target protein, but most often requires the cooperative activity of E3 ligases. Reviewed here are recent structural and functional studies that improve our mechanistic understanding of E1-, E2-, and E3-mediated Ubl conjugation.
Keywords: E1; E2; E3; ligase; ubiquitin; ubiquitin-like protein.
Figures
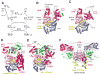
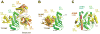
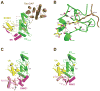
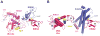
Similar articles
-
Strategies to Trap Enzyme-Substrate Complexes that Mimic Michaelis Intermediates During E3-Mediated Ubiquitin-Like Protein Ligation.Methods Mol Biol. 2018;1844:169-196. doi: 10.1007/978-1-4939-8706-1_12. Methods Mol Biol. 2018. PMID: 30242710 Free PMC article.
-
Binding to E1 and E3 is mutually exclusive for the human autophagy E2 Atg3.Protein Sci. 2013 Dec;22(12):1691-7. doi: 10.1002/pro.2381. Protein Sci. 2013. PMID: 24186333 Free PMC article.
-
NEDD8 and ubiquitin ligation by cullin-RING E3 ligases.Curr Opin Struct Biol. 2021 Apr;67:101-109. doi: 10.1016/j.sbi.2020.10.007. Epub 2020 Nov 5. Curr Opin Struct Biol. 2021. PMID: 33160249 Free PMC article. Review.
-
The ligation systems for ubiquitin and ubiquitin-like proteins.Mol Cells. 1998 Oct 31;8(5):503-12. Mol Cells. 1998. PMID: 9856335 Review.
-
Protein-protein interactions regulate Ubl conjugation.Curr Opin Struct Biol. 2007 Dec;17(6):665-73. doi: 10.1016/j.sbi.2007.09.001. Epub 2007 Oct 15. Curr Opin Struct Biol. 2007. PMID: 17933515
Cited by
-
Expansion of the HSP70 gene family in Tegillarca granosa and expression profiles in response to zinc toxicity.Cell Stress Chaperones. 2024 Feb;29(1):97-112. doi: 10.1016/j.cstres.2024.01.004. Epub 2024 Jan 24. Cell Stress Chaperones. 2024. PMID: 38272254 Free PMC article.
-
Crystal structures of an E1-E2-ubiquitin thioester mimetic reveal molecular mechanisms of transthioesterification.Nat Commun. 2021 Apr 22;12(1):2370. doi: 10.1038/s41467-021-22598-y. Nat Commun. 2021. PMID: 33888705 Free PMC article.
-
The ubiquitin-conjugating enzyme UBE2QL1 coordinates lysophagy in response to endolysosomal damage.EMBO Rep. 2019 Oct 4;20(10):e48014. doi: 10.15252/embr.201948014. Epub 2019 Aug 21. EMBO Rep. 2019. PMID: 31432621 Free PMC article.
-
Whole-Transcriptome Analysis Unveils the Synchronized Activities of Genes for Fructans in Developing Tubers of the Jerusalem Artichoke.Front Plant Sci. 2020 Feb 21;11:101. doi: 10.3389/fpls.2020.00101. eCollection 2020. Front Plant Sci. 2020. PMID: 32153609 Free PMC article.
-
Bacterial Proteasomes.Annu Rev Microbiol. 2015;69:109-27. doi: 10.1146/annurev-micro-091014-104201. Annu Rev Microbiol. 2015. PMID: 26488274 Free PMC article. Review.
References
-
- Aravind L, Koonin EV. The U box is a modified RING finger---a common domain in ubiquitination. Curr Biol. 2000;10:R132–34. - PubMed
-
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–56. - PubMed
-
- Boh BK, Smith PG, Hagen T. Neddylation-induced conformational control regulates cullin RING ligase activity in vivo. J Mol Biol. 2011;409:136–45. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

