Rad51-Rad52 mediated maintenance of centromeric chromatin in Candida albicans
- PMID: 24762765
- PMCID: PMC3998917
- DOI: 10.1371/journal.pgen.1004344
Rad51-Rad52 mediated maintenance of centromeric chromatin in Candida albicans
Abstract
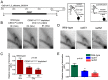
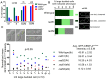
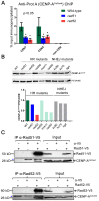
Specification of the centromere location in most eukaryotes is not solely dependent on the DNA sequence. However, the non-genetic determinants of centromere identity are not clearly defined. While multiple mechanisms, individually or in concert, may specify centromeres epigenetically, most studies in this area are focused on a universal factor, a centromere-specific histone H3 variant CENP-A, often considered as the epigenetic determinant of centromere identity. In spite of variable timing of its loading at centromeres across species, a replication coupled early S phase deposition of CENP-A is found in most yeast centromeres. Centromeres are the earliest replicating chromosomal regions in a pathogenic budding yeast Candida albicans. Using a 2-dimensional agarose gel electrophoresis assay, we identify replication origins (ORI7-LI and ORI7-RI) proximal to an early replicating centromere (CEN7) in C. albicans. We show that the replication forks stall at CEN7 in a kinetochore dependent manner and fork stalling is reduced in the absence of the homologous recombination (HR) proteins Rad51 and Rad52. Deletion of ORI7-RI causes a significant reduction in the stalled fork signal and an increased loss rate of the altered chromosome 7. The HR proteins, Rad51 and Rad52, have been shown to play a role in fork restart. Confocal microscopy shows declustered kinetochores in rad51 and rad52 mutants, which are evidence of kinetochore disintegrity. CENP-ACaCse4 levels at centromeres, as determined by chromatin immunoprecipitation (ChIP) experiments, are reduced in absence of Rad51/Rad52 resulting in disruption of the kinetochore structure. Moreover, western blot analysis reveals that delocalized CENP-A molecules in HR mutants degrade in a similar fashion as in other kinetochore mutants described before. Finally, co-immunoprecipitation assays indicate that Rad51 and Rad52 physically interact with CENP-ACaCse4 in vivo. Thus, the HR proteins Rad51 and Rad52 epigenetically maintain centromere functioning by regulating CENP-ACaCse4 levels at the programmed stall sites of early replicating centromeres.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Regulation of mitotic recombination between DNA repeats in centromeres.Nucleic Acids Res. 2017 Nov 2;45(19):11222-11235. doi: 10.1093/nar/gkx763. Nucleic Acids Res. 2017. PMID: 28977643 Free PMC article.
-
A coordinated interdependent protein circuitry stabilizes the kinetochore ensemble to protect CENP-A in the human pathogenic yeast Candida albicans.PLoS Genet. 2012;8(4):e1002661. doi: 10.1371/journal.pgen.1002661. Epub 2012 Apr 19. PLoS Genet. 2012. PMID: 22536162 Free PMC article.
-
Rapid evolution of Cse4p-rich centromeric DNA sequences in closely related pathogenic yeasts, Candida albicans and Candida dubliniensis.Proc Natl Acad Sci U S A. 2008 Dec 16;105(50):19797-802. doi: 10.1073/pnas.0809770105. Epub 2008 Dec 5. Proc Natl Acad Sci U S A. 2008. PMID: 19060206 Free PMC article.
-
The ins and outs of CENP-A: Chromatin dynamics of the centromere-specific histone.Semin Cell Dev Biol. 2023 Feb 15;135:24-34. doi: 10.1016/j.semcdb.2022.04.003. Epub 2022 Apr 11. Semin Cell Dev Biol. 2023. PMID: 35422390 Review.
-
Gross Chromosomal Rearrangement at Centromeres.Biomolecules. 2023 Dec 24;14(1):28. doi: 10.3390/biom14010028. Biomolecules. 2023. PMID: 38254628 Free PMC article. Review.
Cited by
-
Loss of centromere function drives karyotype evolution in closely related Malassezia species.Elife. 2020 Jan 20;9:e53944. doi: 10.7554/eLife.53944. Elife. 2020. PMID: 31958060 Free PMC article.
-
Lessons learned from counting molecules: how to lure CENP-A into the kinetochore.Open Biol. 2014 Dec;4(12):140191. doi: 10.1098/rsob.140191. Open Biol. 2014. PMID: 25500356 Free PMC article. No abstract available.
-
Expansion of human centromeric arrays in cells undergoing break-induced replication.bioRxiv [Preprint]. 2023 Nov 15:2023.11.11.566714. doi: 10.1101/2023.11.11.566714. bioRxiv. 2023. Update in: Cell Rep. 2024 Mar 26;43(3):113851. doi: 10.1016/j.celrep.2024.113851 PMID: 38014305 Free PMC article. Updated. Preprint.
-
Rad52 phosphorylation by Ipl1 and Mps1 contributes to Mps1 kinetochore localization and spindle assembly checkpoint regulation.Proc Natl Acad Sci U S A. 2017 Oct 31;114(44):E9261-E9270. doi: 10.1073/pnas.1705261114. Epub 2017 Oct 16. Proc Natl Acad Sci U S A. 2017. PMID: 29078282 Free PMC article.
-
Orc4 spatiotemporally stabilizes centromeric chromatin.Genome Res. 2021 Apr;31(4):607-621. doi: 10.1101/gr.265900.120. Epub 2021 Jan 29. Genome Res. 2021. PMID: 33514624 Free PMC article.
References
-
- Earnshaw WC, Migeon BR (1985) Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma 92: 290–296. - PubMed
-
- Yamazaki S, Hayano M, Masai H (2013) Replication timing regulation of eukaryotic replicons: Rif1 as a global regulator of replication timing. Trends Genet 29: 449–460. - PubMed
-
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, et al. (2001) Replication dynamics of the yeast genome. Science 294: 115–121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

