Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community
- PMID: 24753569
- PMCID: PMC4020051
- DOI: 10.1073/pnas.1401006111
Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community
Abstract
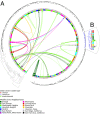
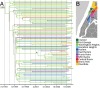
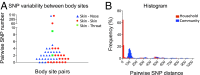
During the last 2 decades, community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) strains have dramatically increased the global burden of S. aureus infections. The pandemic sequence type (ST)8/pulsed-field gel type USA300 is the dominant CA-MRSA clone in the United States, but its evolutionary history and basis for biological success are incompletely understood. Here, we use whole-genome sequencing of 387 ST8 isolates drawn from an epidemiological network of CA-MRSA infections and colonizations in northern Manhattan to explore short-term evolution and transmission patterns. Phylogenetic analysis predicted that USA300 diverged from a most common recent ancestor around 1993. We found evidence for multiple introductions of USA300 and reconstructed the phylogeographic spread of isolates across neighborhoods. Using pair-wise single-nucleotide polymorphism distances as a measure of genetic relatedness between isolates, we observed that most USA300 isolates had become endemic in households, indicating their critical role as reservoirs for transmission and diversification. Using the maximum single-nucleotide polymorphism variability of isolates from within households as a threshold, we identified several possible transmission networks beyond households. Our study also revealed the evolution of a fluoroquinolone-resistant subpopulation in the mid-1990s and its subsequent expansion at a time of high-frequency outpatient antibiotic use. This high-resolution phylogenetic analysis of ST8 has documented the genomic changes associated with USA300 evolution and how some of its recent evolution has been shaped by antibiotic use. By integrating whole-genome sequencing with detailed epidemiological analyses, our study provides an important framework for delineating the full diversity and spread of USA300 and other emerging pathogens in large urban community populations.
Keywords: CC8; drug resistance; genomics; phylogeny.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Demography and Intercontinental Spread of the USA300 Community-Acquired Methicillin-Resistant Staphylococcus aureus Lineage.mBio. 2016 Feb 16;7(1):e02183-15. doi: 10.1128/mBio.02183-15. mBio. 2016. PMID: 26884428 Free PMC article.
-
Transmission and microevolution of USA300 MRSA in U.S. households: evidence from whole-genome sequencing.mBio. 2015 Mar 10;6(2):e00054. doi: 10.1128/mBio.00054-15. mBio. 2015. PMID: 25759497 Free PMC article.
-
Evolution and Global Transmission of a Multidrug-Resistant, Community-Associated Methicillin-Resistant Staphylococcus aureus Lineage from the Indian Subcontinent.mBio. 2019 Nov 26;10(6):e01105-19. doi: 10.1128/mBio.01105-19. mBio. 2019. PMID: 31772058 Free PMC article.
-
Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA).Curr Opin Microbiol. 2012 Oct;15(5):588-95. doi: 10.1016/j.mib.2012.08.003. Epub 2012 Oct 5. Curr Opin Microbiol. 2012. PMID: 23044073 Review.
-
Virulence strategies of the dominant USA300 lineage of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA).FEMS Immunol Med Microbiol. 2012 Jun;65(1):5-22. doi: 10.1111/j.1574-695X.2012.00937.x. Epub 2012 Mar 5. FEMS Immunol Med Microbiol. 2012. PMID: 22309135 Free PMC article. Review.
Cited by
-
Emergent and evolving antimicrobial resistance cassettes in community-associated fusidic acid and meticillin-resistant Staphylococcus aureus.Int J Antimicrob Agents. 2015 May;45(5):477-84. doi: 10.1016/j.ijantimicag.2015.01.009. Epub 2015 Feb 19. Int J Antimicrob Agents. 2015. PMID: 25769787 Free PMC article.
-
Global Spread of the Community-Associated Methicillin-Resistant Staphylococcus aureus USA300 Latin American Variant.J Infect Dis. 2016 Nov 15;214(10):1609-1610. doi: 10.1093/infdis/jiw418. Epub 2016 Sep 9. J Infect Dis. 2016. PMID: 27613776 Free PMC article. No abstract available.
-
A Complete Genome Screening Program of Clinical Methicillin-Resistant Staphylococcus aureus Isolates Identifies the Origin and Progression of a Neonatal Intensive Care Unit Outbreak.J Clin Microbiol. 2019 Nov 22;57(12):e01261-19. doi: 10.1128/JCM.01261-19. Print 2019 Dec. J Clin Microbiol. 2019. PMID: 31578260 Free PMC article.
-
The Microevolution and Epidemiology of Staphylococcus aureus Colonization during Atopic Eczema Disease Flare.J Invest Dermatol. 2018 Feb;138(2):336-343. doi: 10.1016/j.jid.2017.09.023. Epub 2017 Sep 23. J Invest Dermatol. 2018. PMID: 28951239 Free PMC article.
-
The epidemiology and transmission of methicillin-resistant Staphylococcus aureus in the community in Singapore: study protocol for a longitudinal household study.BMC Infect Dis. 2017 Oct 11;17(1):678. doi: 10.1186/s12879-017-2793-y. BMC Infect Dis. 2017. PMID: 29020940 Free PMC article.
References
-
- Fridkin SK, et al. Active Bacterial Core Surveillance Program of the Emerging Infections Program Network Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med. 2005;352(14):1436–1444. - PubMed
-
- Moran GJ, et al. EMERGEncy ID Net Study Group Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355(7):666–674. - PubMed
-
- Talan DA, et al. EMERGEncy ID Net Study Group Comparison of Staphylococcus aureus from skin and soft-tissue infections in US emergency department patients, 2004 and 2008. Clin Infect Dis. 2011;53(2):144–149. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

