E2-mediated small ubiquitin-like modifier (SUMO) modification of thymine DNA glycosylase is efficient but not selective for the enzyme-product complex
- PMID: 24753249
- PMCID: PMC4140935
- DOI: 10.1074/jbc.M114.572081
E2-mediated small ubiquitin-like modifier (SUMO) modification of thymine DNA glycosylase is efficient but not selective for the enzyme-product complex
Abstract
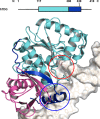
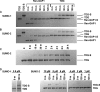
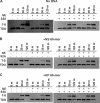
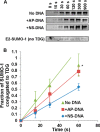
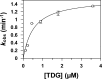
Thymine DNA glycosylase (TDG) initiates the repair of G·T mismatches that arise by deamination of 5-methylcytosine (mC), and it excises 5-formylcytosine and 5-carboxylcytosine, oxidized forms of mC. TDG functions in active DNA demethylation and is essential for embryonic development. TDG forms a tight enzyme-product complex with abasic DNA, which severely impedes enzymatic turnover. Modification of TDG by small ubiquitin-like modifier (SUMO) proteins weakens its binding to abasic DNA. It was proposed that sumoylation of product-bound TDG regulates product release, with SUMO conjugation and deconjugation needed for each catalytic cycle, but this model remains unsubstantiated. We examined the efficiency and specificity of TDG sumoylation using in vitro assays with purified E1 and E2 enzymes, finding that TDG is modified efficiently by SUMO-1 and SUMO-2. Remarkably, we observed similar modification rates for free TDG and TDG bound to abasic or undamaged DNA. To examine the conjugation step directly, we determined modification rates (kobs) using preformed E2∼SUMO-1 thioester. The hyperbolic dependence of kobs on TDG concentration gives kmax = 1.6 min(-1) and K1/2 = 0.55 μM, suggesting that E2∼SUMO-1 has higher affinity for TDG than for the SUMO targets RanGAP1 and p53 (peptide). Whereas sumoylation substantially weakens TDG binding to DNA, TDG∼SUMO-1 still binds relatively tightly to AP-DNA (Kd ∼50 nM). Although E2∼SUMO-1 exhibits no specificity for product-bound TDG, the relatively high conjugation efficiency raises the possibility that E2-mediated sumoylation could stimulate product release in vivo. This and other implications for the biological role and mechanism of TDG sumoylation are discussed.
Keywords: Base Excision Repair (BER); DNA Methylation; DNA Repair; Enzyme Turnover; Post-translational Modification (PTM); Small Ubiquitin-like Modifier (SUMO); Sumoylation; Ubiquitin-conjugating Enzyme (E2 Enzyme).
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
Characterizing Requirements for Small Ubiquitin-like Modifier (SUMO) Modification and Binding on Base Excision Repair Activity of Thymine-DNA Glycosylase in Vivo.J Biol Chem. 2016 Apr 22;291(17):9014-24. doi: 10.1074/jbc.M115.706325. Epub 2016 Feb 25. J Biol Chem. 2016. PMID: 26917720 Free PMC article.
-
Defining the impact of sumoylation on substrate binding and catalysis by thymine DNA glycosylase.Nucleic Acids Res. 2018 Jun 1;46(10):5159-5170. doi: 10.1093/nar/gky278. Nucleic Acids Res. 2018. PMID: 29660017 Free PMC article.
-
Sumoylation of thymine DNA glycosylase impairs productive binding to substrate sites in DNA.J Biol Chem. 2024 Nov;300(11):107902. doi: 10.1016/j.jbc.2024.107902. Epub 2024 Oct 18. J Biol Chem. 2024. PMID: 39426728 Free PMC article.
-
Molecular mechanisms in SUMO conjugation.Biochem Soc Trans. 2020 Feb 28;48(1):123-135. doi: 10.1042/BST20190357. Biochem Soc Trans. 2020. PMID: 31872228 Review.
-
Mechanisms and functions of SUMOylation in health and disease: a review focusing on immune cells.J Biomed Sci. 2024 Jan 27;31(1):16. doi: 10.1186/s12929-024-01003-y. J Biomed Sci. 2024. PMID: 38280996 Free PMC article. Review.
Cited by
-
Progress of small ubiquitin-related modifiers in kidney diseases.Chin Med J (Engl). 2019 Feb;132(4):466-473. doi: 10.1097/CM9.0000000000000094. Chin Med J (Engl). 2019. PMID: 30707172 Free PMC article. Review.
-
Characterizing Requirements for Small Ubiquitin-like Modifier (SUMO) Modification and Binding on Base Excision Repair Activity of Thymine-DNA Glycosylase in Vivo.J Biol Chem. 2016 Apr 22;291(17):9014-24. doi: 10.1074/jbc.M115.706325. Epub 2016 Feb 25. J Biol Chem. 2016. PMID: 26917720 Free PMC article.
-
Role of Base Excision "Repair" Enzymes in Erasing Epigenetic Marks from DNA.Chem Rev. 2016 Oct 26;116(20):12711-12729. doi: 10.1021/acs.chemrev.6b00191. Epub 2016 Aug 8. Chem Rev. 2016. PMID: 27501078 Free PMC article. Review.
-
Defining the impact of sumoylation on substrate binding and catalysis by thymine DNA glycosylase.Nucleic Acids Res. 2018 Jun 1;46(10):5159-5170. doi: 10.1093/nar/gky278. Nucleic Acids Res. 2018. PMID: 29660017 Free PMC article.
-
Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.Nucleic Acids Res. 2015 Oct 30;43(19):9541-52. doi: 10.1093/nar/gkv890. Epub 2015 Sep 10. Nucleic Acids Res. 2015. PMID: 26358812 Free PMC article.
References
-
- Neddermann P., Jiricny J. (1993) The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J. Biol. Chem. 268, 21218–21224 - PubMed
-
- Neddermann P., Gallinari P., Lettieri T., Schmid D., Truong O., Hsuan J. J., Wiebauer K., Jiricny J. (1996) Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J. Biol. Chem. 271, 12767–12774 - PubMed
-
- Cortellino S., Xu J., Sannai M., Moore R., Caretti E., Cigliano A., Le Coz M., Devarajan K., Wessels A., Soprano D., Abramowitz L. K., Bartolomei M. S., Rambow F., Bassi M. R., Bruno T., Fanciulli M., Renner C., Klein-Szanto A. J., Matsumoto Y., Kobi D., Davidson I., Alberti C., Larue L., Bellacosa A. (2011) Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146, 67–79 - PMC - PubMed
-
- Cortázar D., Kunz C., Selfridge J., Lettieri T., Saito Y., MacDougall E., Wirz A., Schuermann D., Jacobs A. L., Siegrist F., Steinacher R., Jiricny J., Bird A., Schär P. (2011) Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 470, 419–423 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

