A conserved RNA polymerase III promoter required for gammaherpesvirus TMER transcription and microRNA processing
- PMID: 24747015
- PMCID: PMC4544698
- DOI: 10.1016/j.gene.2014.04.026
A conserved RNA polymerase III promoter required for gammaherpesvirus TMER transcription and microRNA processing
Abstract
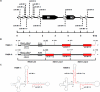
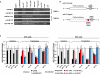
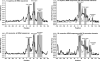
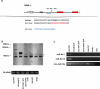
Canonical RNA polymerase III (pol III) type 2 promoters contain a single A and B box and are well documented for their role in tRNA and SINE transcription in eukaryotic cells. The genome of Murid herpesvirus 4 (MuHV-4) contains eight polycistronic tRNA-microRNA encoded RNA (TMER) genes that are transcribed from a RNA pol III type 2-like promoter containing triplicated A box elements. Here, we demonstrate that the triplicated A box sequences are required in their entirety to produce functional MuHV-4 miRNAs. We also identify that these RNA pol III type 2-like promoters are conserved in eukaryotic genomes. Human and mouse predicted tRNA genes containing these promoters also show enrichment of alternative RNA pol III transcription termination sequences and are predicted to give rise to longer tRNA primary transcripts.
Keywords: A box; Gammaherpesvirus 68; Type 2 promoter; tRNA.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters.RNA. 2010 Jan;16(1):170-85. doi: 10.1261/rna.1873910. Epub 2009 Nov 30. RNA. 2010. PMID: 19948768 Free PMC article.
-
Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts.J Virol. 2010 Oct;84(19):10344-53. doi: 10.1128/JVI.00707-10. Epub 2010 Jul 21. J Virol. 2010. PMID: 20660200 Free PMC article.
-
Lytic Infection with Murine Gammaherpesvirus 68 Activates Host and Viral RNA Polymerase III Promoters and Enhances Noncoding RNA Expression.J Virol. 2021 Jun 24;95(14):e0007921. doi: 10.1128/JVI.00079-21. Epub 2021 Jun 24. J Virol. 2021. PMID: 33910955 Free PMC article.
-
Interactions between RNAP III transcription machinery and tRNA processing factors.Biochim Biophys Acta Gene Regul Mech. 2018 Apr;1861(4):354-360. doi: 10.1016/j.bbagrm.2018.02.003. Epub 2018 Feb 9. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 29428193 Review.
-
RNA polymerase III. Genes, factors and transcriptional specificity.Eur J Biochem. 1993 Feb 15;212(1):1-11. doi: 10.1111/j.1432-1033.1993.tb17626.x. Eur J Biochem. 1993. PMID: 8444147 Review.
Cited by
-
MicroRNA-Regulated Proinflammatory Cytokines in Sarcopenia.Mediators Inflamm. 2016;2016:1438686. doi: 10.1155/2016/1438686. Epub 2016 Jun 13. Mediators Inflamm. 2016. PMID: 27382188 Free PMC article. Review.
-
DUSP11 activity on triphosphorylated transcripts promotes Argonaute association with noncanonical viral microRNAs and regulates steady-state levels of cellular noncoding RNAs.Genes Dev. 2016 Sep 15;30(18):2076-2092. doi: 10.1101/gad.282616.116. Genes Dev. 2016. PMID: 27798849 Free PMC article.
-
DUSP11 - An RNA phosphatase that regulates host and viral non-coding RNAs in mammalian cells.RNA Biol. 2017 Nov 2;14(11):1457-1465. doi: 10.1080/15476286.2017.1306169. Epub 2017 Apr 17. RNA Biol. 2017. PMID: 28296624 Free PMC article.
-
Murine Gammaherpesvirus 68 ORF45 Stimulates B2 Retrotransposon and Pre-tRNA Activation in a Manner Dependent on Mitogen-Activated Protein Kinase (MAPK) Signaling.Microbiol Spectr. 2023 Feb 8;11(2):e0017223. doi: 10.1128/spectrum.00172-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 36752632 Free PMC article.
-
Identification of a polyomavirus microRNA highly expressed in tumors.Virology. 2015 Feb;476:43-53. doi: 10.1016/j.virol.2014.11.021. Epub 2014 Dec 13. Virology. 2015. PMID: 25514573 Free PMC article.
References
-
- Barton E, Mandal P, Speck S. Pathogenesis and host control of gammaherpesviruses: lessons from the mouse. Annu. Rev. Immunol. 2011;29:351–397. - PubMed
-
- Blaskovic D, Stanceková M, Svobodová J, Mistríková J. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 1980;24:468. - PubMed
-
- Bogenhagen DF, Brown DD. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981;24:261–270. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

