CTCF binding to the first intron of the major immediate early (MIE) gene of human cytomegalovirus (HCMV) negatively regulates MIE gene expression and HCMV replication
- PMID: 24741094
- PMCID: PMC4054410
- DOI: 10.1128/JVI.00845-14
CTCF binding to the first intron of the major immediate early (MIE) gene of human cytomegalovirus (HCMV) negatively regulates MIE gene expression and HCMV replication
Abstract
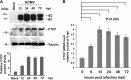
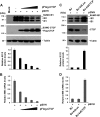
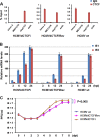
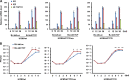
Human cytomegalovirus (HCMV) gene expression during infection is highly regulated, with sequential expression of immediate-early (IE), early (E), and late (L) gene transcripts. To explore the potential role of chromatin regulatory factors that may regulate HCMV gene expression and DNA replication, we investigated the interaction of HCMV with the cellular chromatin-organizing factor CTCF. Here, we show that HCMV-infected cells produce higher levels of CTCF mRNA and protein at early stages of infection. We also show that CTCF depletion by short hairpin RNA results in an increase in major IE (MIE) and E gene expression and an about 50-fold increase in HCMV particle production. We identified a DNA sequence (TTAACGGTGGAGGGCAGTGT) in the first intron (intron A) of the MIE gene that interacts directly with CTCF. Deletion of this CTCF-binding site led to an increase in MIE gene expression in both transient-transfection and infection assays. Deletion of the CTCF-binding site in the HCMV bacterial artificial chromosome plasmid genome resulted in an about 10-fold increase in the rate of viral replication relative to either wild-type or revertant HCMV. The CTCF-binding site deletion had no detectable effect on MIE gene-splicing regulation, nor did CTCF knockdown or overexpression of CTCF alter the ratio of IE1 to IE2. Therefore, CTCF binds to DNA within the MIE gene at the position of the first intron to affect RNA polymerase II function during the early stages of viral transcription. Finally, the CTCF-binding sequence in CMV is evolutionarily conserved, as a similar sequence in murine CMV (MCMV) intron A was found to interact with CTCF and similarly function in the repression of MCMV MIE gene expression mediated by CTCF.
Importance: Our findings that CTCF binds to intron A of the cytomegalovirus (CMV) major immediate-early (MIE) gene and functions to repress MIE gene expression and viral replication are highly significant. For the first time, a chromatin-organizing factor, CTCF, has been found to facilitate human CMV gene expression, which affects viral replication. We also identified a CTCF-binding motif in the first intron (also called intron A) that directly binds to CTCF and is required for CTCF to repress MIE gene expression. Finally, we show that the CTCF-binding motif is conserved in CMV because a similar DNA sequence was found in murine CMV (MCMV) that is required for CTCF to bind to MCMV MIE gene to repress MCMV MIE gene expression.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
The 5' Untranslated Region of the Major Immediate Early mRNA Is Necessary for Efficient Human Cytomegalovirus Replication.J Virol. 2018 Mar 14;92(7):e02128-17. doi: 10.1128/JVI.02128-17. Print 2018 Apr 1. J Virol. 2018. PMID: 29343581 Free PMC article.
-
Two Polypyrimidine Tracts in Intron 4 of the Major Immediate Early Gene Are Critical for Gene Expression Switching from IE1 to IE2 and for Replication of Human Cytomegalovirus.J Virol. 2016 Jul 27;90(16):7339-7349. doi: 10.1128/JVI.00837-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27252533 Free PMC article.
-
Multiple Transcripts Encode Full-Length Human Cytomegalovirus IE1 and IE2 Proteins during Lytic Infection.J Virol. 2016 Sep 12;90(19):8855-65. doi: 10.1128/JVI.00741-16. Print 2016 Oct 1. J Virol. 2016. PMID: 27466417 Free PMC article.
-
Loopy virus or controlled contortionist? 3D regulation of HCMV gene expression by CTCF-driven chromatin interactions.J Virol. 2024 Oct 22;98(10):e0114824. doi: 10.1128/jvi.01148-24. Epub 2024 Aug 30. J Virol. 2024. PMID: 39212383 Review.
-
Bright and Early: Inhibiting Human Cytomegalovirus by Targeting Major Immediate-Early Gene Expression or Protein Function.Viruses. 2020 Jan 16;12(1):110. doi: 10.3390/v12010110. Viruses. 2020. PMID: 31963209 Free PMC article. Review.
Cited by
-
Complex Interactions between Cohesin and CTCF in Regulation of Kaposi's Sarcoma-Associated Herpesvirus Lytic Transcription.J Virol. 2020 Jan 6;94(2):e01279-19. doi: 10.1128/JVI.01279-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31666380 Free PMC article.
-
Functional annotation of human cytomegalovirus gene products: an update.Front Microbiol. 2014 May 19;5:218. doi: 10.3389/fmicb.2014.00218. eCollection 2014. Front Microbiol. 2014. PMID: 24904534 Free PMC article. Review.
-
The three-dimensional structure of the EBV genome plays a crucial role in regulating viral gene expression in EBVaGC.Nucleic Acids Res. 2023 Dec 11;51(22):12092-12110. doi: 10.1093/nar/gkad936. Nucleic Acids Res. 2023. PMID: 37889078 Free PMC article.
-
The 5' Untranslated Region of the Major Immediate Early mRNA Is Necessary for Efficient Human Cytomegalovirus Replication.J Virol. 2018 Mar 14;92(7):e02128-17. doi: 10.1128/JVI.02128-17. Print 2018 Apr 1. J Virol. 2018. PMID: 29343581 Free PMC article.
-
CCCTC-Binding Factor Acts as a Heterochromatin Barrier on Herpes Simplex Viral Latent Chromatin and Contributes to Poised Latent Infection.mBio. 2018 Feb 6;9(1):e02372-17. doi: 10.1128/mBio.02372-17. mBio. 2018. PMID: 29437926 Free PMC article.
References
-
- Mocarski ES, Jr, Shenk T, Pass RF. 2006. Cytomegaloviruses, 5th ed. Lippincott Williams &Wilkins, Philadelphia, PA
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

